Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks - PubMed (original) (raw)
Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks
François Aymard et al. Nat Struct Mol Biol. 2014 Apr.
Abstract
Although both homologous recombination (HR) and nonhomologous end joining can repair DNA double-strand breaks (DSBs), the mechanisms by which one of these pathways is chosen over the other remain unclear. Here we show that transcriptionally active chromatin is preferentially repaired by HR. Using chromatin immunoprecipitation-sequencing (ChIP-seq) to analyze repair of multiple DSBs induced throughout the human genome, we identify an HR-prone subset of DSBs that recruit the HR protein RAD51, undergo resection and rely on RAD51 for efficient repair. These DSBs are located in actively transcribed genes and are targeted to HR repair via the transcription elongation-associated mark trimethylated histone H3 K36. Concordantly, depletion of SETD2, the main H3 K36 trimethyltransferase, severely impedes HR at such DSBs. Our study thereby demonstrates a primary role in DSB repair of the chromatin context in which a break occurs.
Figures
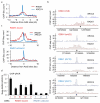
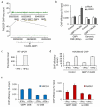
Figure 1. A subset of AsiSI-induced DSBs recruits RAD51
a. ChIP-seq analyses in DIvA cells after 4OHT treatment (4h), using anti XRCC4 or anti-RAD51 antibodies. The averaged XRCC4 (blue) and RAD51 (red) signals, over a 20kb region flanking annotated AsiSI sites, are shown. b. The profiles of XRCC4 (blue) and RAD51 (red) around four selected AsiSI sites (indicated by arrows) are shown. c. Averaged XRCC4 (blue) and RAD51 (red) signals over 20kb windows and centered at the AsiSI site, are shown for each categories (RAD51-bound or RAD51-unbound subsets). d. ChIP against XRCC4 and RAD51 in 4OHT-treated DIvA cells, analyzed by qPCR. The ratios between the signals observed for XRCC4 and RAD51 are presented for eight AsiSI-induced DSBs, either bound or not by RAD51 in ChIP-seq experiments (RAD51-bound DSBs are labeled with roman numerals, while RAD51-unbound DSBs are labeled with arabic numerals). Mean and s.e.m (technical replicate, n=4) of a representative experiment are shown.
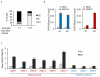
Figure 2. RAD51 recruitment at RAD51-bound DSB occurs mainly in G2
a. Cell cycle distributions as measured by FACS in synchronized DIvA cells b. ChIP using XRCC4 (blue, left panels) or RAD51 (red, right panels) antibodies in synchronized G1 and G2 DIvA cells. Enrichments in XRCC4 and RAD51 were respectively measured by qPCR at 80 bp and 800 bp from the DSB-I (from our RAD51-bound subset). Mean and s.e.m of a representative experiment is shown. c. The RAD51/XRCC4 ratio obtained by ChIP-qPCR was calculated for various DSBs from the RAD51-unbound subset (named with arabic numerals, in blue) and from the RAD51-bound subset (named with roman numerals, in red) both in G1 and G2 cells. RAD51 recruitment at DSBs occurs preferentially in G2, and on a specific subset of DSBs. Mean and s.e.m (n=4 technical replicates) of a representative experiment are shown.
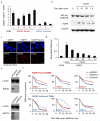
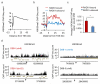
Figure 3. RAD51-bound AsiSI-DSBs are resected and repaired by a RAD51-dependent pathway
a. ssDNA arising through resection was assessed at each of the eight previously analyzed DSBs. Pull down efficiency was measured by RT-qPCR. Ratios between treated and untreated cells are presented. Mean and s.e.m (n=3, technical replicates) of a representative experiment are shown. b. Western blot analysis of AID-DIvA stably transfected cells induced by 4OHT for 4h and subsequently treated (or not) with auxin for the indicated time. Uncropped images are presented Supplementary Fig.8c. The presence of γH2AX foci was monitored by immunofluorescence in untreated cells, in 4OHT-induced cells (4h), and in 4OHT-induced cells further incubated with auxin (2h). d. Cleavage assay in AID-DIvA cells, treated as indicated, followed by RT-qPCR close to a cleaved AsiSI site. Normalized pull down efficiencies from a representative experiment are shown. e. Western blot analyses of AID-DIvA cells transfected with a control siRNA (Control), siRNA against RAD51 (left panel) or against XRCC4 (right panel). Uncropped images are presented Supplementary Fig.8. f. Cleavage assay in AID-DIvA cells transfected with control, RAD51, or XRCC4 siRNAs. Immunoprecipitated DNA was analyzed close to six DSBs, either RAD51-unbound (indicated in blue, upper panel) or RAD51-bound (indicated in red, lower panel). The percentage of sites that remain broken for each DSB after the indicated times of auxin treatment are presented. A representative experiment is shown.
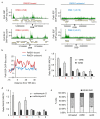
Figure 4. RAD51-bound DSBs lie within transcribed units
a. PolII-S2P ChIP-seq in untreated DIvA cells. PolII-S2P enrichment is shown around the four AsiSI-induced DSBs presented Fig. 1b. RAD51-bound DSBs are indicated in red and RAD51-unbound DSBs in blue. Positions are in bp. b. The genes associated with each DSB from the RAD51-bound and RAD51-unbound subsets were used to calculate the average PolII-S2P enrichments around the transcription start site. c. XRCC4 and RAD51 ChIP in 4OHT-treated DIvA incubated with DRB (or not). The RAD51/XRCC4 ratios for three RAD51-bound DSBs (DSB-I, -II and -III), one RAD51-unbound DSB (DSB-1), and two DSBs located far from any gene (DSB-5, and -6) are shown. Mean and s.e.m (n=4, technical replicates) of a representative experiment are shown d. Same as c, except that cells were treated with Actinomycin D or not. e. Cell cycle distribution measured by EdU and PI staining and FACS analysis, in untreated, DRB, or Actinomycin D treated DIvA cells, as indicated. Mean and s.e.m of percent of cells in each phase are shown (n=3, biological replicates).
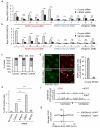
Figure 5. Transcriptional activation of an unexpressed gene leads to repair by HR
a. PolII-S2P profile obtained in DIvA cells is shown on the GBP cluster of gene, located on chromosome 1. A TALEN pair was ordered to specifically induce a DSB in the GBP1 gene, part of the GBP cluster located on chromosome 1. b. γH2AX ChIP in DIvA cells transfected either with pCDNA3 or with the GBP1-TALEN plasmids. Enrichment was measured by qPCR at 80bp from the expected DSB, or on a control genomic region. ChIP efficiency (expressed as % of input) of a representative experiment is shown. c. Levels of GBP1 mRNA measured by reverse transcription followed by qPCR, in DIvA cells treated (or not) with IFN-γ as indicated. cDNA levels were normalized to ribosomal protein P0 cDNA levels. d. H3K36me3 ChIP in control or TALEN transfected cells subjected (or not) to an IFN-γ treatment. H3K36me3 levels were analyzed by qPCR on the GBP1 gene. Mean and s.e.m (n=4, technical replicates) of a representative experiment are shown. e. XRCC4 (blue) and RAD51 (red) ChIP in GBP1-TALEN transfected cells, treated or not with γ-IFN. Enrichments of XRCC4 and RAD51 were measured by qPCR respectively at 80bp and 800bp from the TALEN-induced DSB. Mean and s.e.m (n=4, technical replicates) of a representative experiment are shown.
Figure 6. H3K36 me3 enrichment on chromatin correlates with the use of HR
a. H3K36me3 ChIP-seq in untreated DIvA cells. The average H3K36me3 signal was calculated around transcriptional start site (TSS) of all annotated genes (RefSeq, hg18). b. The genes associated with each DSB from the RAD51-bound and RAD51-unbound subsets (respectively red and blue lines) were used to calculate the average H3K36me3 enrichments around the TSS. c. H3K36me3 average signal calculated over a 4kb window centered on the DSB, for each category are shown, together with the s.e.m. * p<0.05 (Mann Whitney).d. Profiles of H3K36me3 in undamaged DIvA cells at two RAD51-bound DSBs (DSB-I and DSB-II), one DSB identified in our RAD51-unbound subset (DSB-1), and a DSB located far from any gene (DSB-5).
Figure 7. The H3K36me3-LEDGF axis targets RAD51 at DSBs induced in active genes
a. XRCC4 and RAD51 ChIP in 4OHT-treated DIvA cells transfected with control or LEDGF siRNAs. RAD51/XRCC4 ratios were analyzed by RT-qPCR around six RAD51-bound DSBs (DSB-I to-VI), four RAD51-unbound DSBs (DSB-1 to-4) and two DSBs far from any gene (DSB-5 and -6). * p<0.05; ** p<0.01; *** p<0.005; ns non-significant (unpaired Student t-test, two sided). b. Same as in c, except that a siRNA against SETD2 was used. c. Cell cycle distribution measured by EdU and PI staining and FACS analysis, in DIvA cells transfected with siRNA, as indicated. Mean and s.e.m of percent of cells in each phase are shown (n=3, biological replicates). d. U2OS cells, transfected with siRNAs as indicated, were laser micro-irradiated and stained for cyclinA, γH2AX (red) and RAD51 (green). Percentages of cyclin A positive cells, that exhibit RAD51 recruitment at the laser line are shown. e. RG37-GFP-HR reporter cells were transfected with siRNAs as indicated and with a vector expressing I-SceI or not. GFP positive cells were measured by flow cytometry, and expressed relative to the amount of GFP positive cells in control siRNA transfected cells. Mean and s.e.m of independent experiments are shown (n=4, biological replicates). * p<0.05; ** p<0.01; *** p<0.005 (unpaired Student t-test, two sided).f. H3K36me3 ChIP-seq in untreated and 4OHT treated DIvA cells. Profiles obtained around the RAD51-bound DSB-I (indicated by an arrow) are shown. Positions are in bp. g. H3K36me3 signals obtained by ChIP-seq were averaged around all cleaved AsiSI sites.
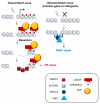
Figure 8. Targeting of HR at transcriptionally active loci
The trimethylation of histone H3 on the lysine 36 (H3K36me3), a mark associated with transcriptional elongation, would be recognized by LEDGF, itself interacting with CtIP, thus bringing to the site of break a resection promoting factor. This would subsequently trigger RAD51 binding and HR repair on actively transcribed genes. DSBs occurring within inactive genes or intergenic regions would be unable to recruit a resection factor thus leading to repair by NHEJ.
Similar articles
- DNA double strand break repair pathway choice: a chromatin based decision?
Clouaire T, Legube G. Clouaire T, et al. Nucleus. 2015;6(2):107-13. doi: 10.1080/19491034.2015.1010946. Epub 2015 Feb 12. Nucleus. 2015. PMID: 25675367 Free PMC article. - The SETD2 Methyltransferase Supports Productive HPV31 Replication through the LEDGF/CtIP/Rad51 Pathway.
Mac M, DeVico BM, Raspanti SM, Moody CA. Mac M, et al. J Virol. 2023 May 31;97(5):e0020123. doi: 10.1128/jvi.00201-23. Epub 2023 May 8. J Virol. 2023. PMID: 37154769 Free PMC article. - Rad51 recruitment and exclusion of non-homologous end joining during homologous recombination at a Tus/Ter mammalian replication fork barrier.
Willis NA, Panday A, Duffey EE, Scully R. Willis NA, et al. PLoS Genet. 2018 Jul 19;14(7):e1007486. doi: 10.1371/journal.pgen.1007486. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30024881 Free PMC article. - Repair of strand breaks by homologous recombination.
Jasin M, Rothstein R. Jasin M, et al. Cold Spring Harb Perspect Biol. 2013 Nov 1;5(11):a012740. doi: 10.1101/cshperspect.a012740. Cold Spring Harb Perspect Biol. 2013. PMID: 24097900 Free PMC article. Review. - The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice.
Frigerio C, Di Nisio E, Galli M, Colombo CV, Negri R, Clerici M. Frigerio C, et al. Int J Mol Sci. 2023 Feb 7;24(4):3248. doi: 10.3390/ijms24043248. Int J Mol Sci. 2023. PMID: 36834658 Free PMC article. Review.
Cited by
- CtIP-dependent nascent RNA expression flanking DNA breaks guides the choice of DNA repair pathway.
Gómez-Cabello D, Pappas G, Aguilar-Morante D, Dinant C, Bartek J. Gómez-Cabello D, et al. Nat Commun. 2022 Sep 9;13(1):5303. doi: 10.1038/s41467-022-33027-z. Nat Commun. 2022. PMID: 36085345 Free PMC article. - TRIM29 regulates the assembly of DNA repair proteins into damaged chromatin.
Masuda Y, Takahashi H, Sato S, Tomomori-Sato C, Saraf A, Washburn MP, Florens L, Conaway RC, Conaway JW, Hatakeyama S. Masuda Y, et al. Nat Commun. 2015 Jun 22;6:7299. doi: 10.1038/ncomms8299. Nat Commun. 2015. PMID: 26095369 - H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex.
Saredi G, Huang H, Hammond CM, Alabert C, Bekker-Jensen S, Forne I, Reverón-Gómez N, Foster BM, Mlejnkova L, Bartke T, Cejka P, Mailand N, Imhof A, Patel DJ, Groth A. Saredi G, et al. Nature. 2016 Jun 30;534(7609):714-718. doi: 10.1038/nature18312. Epub 2016 Jun 22. Nature. 2016. PMID: 27338793 Free PMC article. - Transcriptional regulation and chromatin dynamics at DNA double-strand breaks.
Min S, Ji JH, Heo Y, Cho H. Min S, et al. Exp Mol Med. 2022 Oct;54(10):1705-1712. doi: 10.1038/s12276-022-00862-5. Epub 2022 Oct 13. Exp Mol Med. 2022. PMID: 36229590 Free PMC article. Review. - Epigenomic Analysis of RAD51 ChIP-seq Data Reveals _cis_-regulatory Elements Associated with Autophagy in Cancer Cell Lines.
Kang K, Choi Y, Moon H, You C, Seo M, Kwon G, Yun J, Beck B, Kang K. Kang K, et al. Cancers (Basel). 2021 May 22;13(11):2547. doi: 10.3390/cancers13112547. Cancers (Basel). 2021. PMID: 34067336 Free PMC article.
References
- Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 092096/WT_/Wellcome Trust/United Kingdom
- C6/A11226/CRUK_/Cancer Research UK/United Kingdom
- 268536/ERC_/European Research Council/International
- A11224/CRUK_/Cancer Research UK/United Kingdom
- 086861/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- F31 CA260794/CA/NCI NIH HHS/United States
- 11224/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials