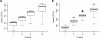FreeContact: fast and free software for protein contact prediction from residue co-evolution - PubMed (original) (raw)
FreeContact: fast and free software for protein contact prediction from residue co-evolution
László Kaján et al. BMC Bioinformatics. 2014.
Abstract
Background: 20 years of improved technology and growing sequences now renders residue-residue contact constraints in large protein families through correlated mutations accurate enough to drive de novo predictions of protein three-dimensional structure. The method EVfold broke new ground using mean-field Direct Coupling Analysis (EVfold-mfDCA); the method PSICOV applied a related concept by estimating a sparse inverse covariance matrix. Both methods (EVfold-mfDCA and PSICOV) are publicly available, but both require too much CPU time for interactive applications. On top, EVfold-mfDCA depends on proprietary software.
Results: Here, we present FreeContact, a fast, open source implementation of EVfold-mfDCA and PSICOV. On a test set of 140 proteins, FreeContact was almost eight times faster than PSICOV without decreasing prediction performance. The EVfold-mfDCA implementation of FreeContact was over 220 times faster than PSICOV with negligible performance decrease. EVfold-mfDCA was unavailable for testing due to its dependency on proprietary software. FreeContact is implemented as the free C++ library "libfreecontact", complete with command line tool "freecontact", as well as Perl and Python modules. All components are available as Debian packages. FreeContact supports the BioXSD format for interoperability.
Conclusions: FreeContact provides the opportunity to compute reliable contact predictions in any environment (desktop or cloud).
Figures
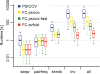
Figure 1
Runtimes for FreeContact. We measured the runtime (logarithmic y-axis) for different program components (x-axis) on a single thread. The program components were: “seqw” – sequence weighting; “pairfreq” – pairwise residue frequencies; “shrink” – shrinking of covariance matrix; “inv” – sparse inverse covariance estimation/covariance matrix inversion. The different colors distinguish: the original PSICOV implementation (blue), our acceleration of PSICOV (FC.psicov, yellow), our acceleration of the faster PSICOV version “sensible default” (FC.psicov-fast, green), and our implementation of EVfold-mfDCA (FC.evfold, red). The whiskers on the box plots show the most extreme data point that is less than 1.5-times the interquartile range from the box. Outliers are not shown. Total runtime of all methods tested is dominated by the sparse inverse covariance estimation/covariance matrix inversion component.
Figure 2
Speedup using multiple threads. A: Sequence weighting. Speed is calculated as: proteins in alignment2 length of target protein/runtime. B: Pairwise residue frequency calculation. Speed is calculated as: proteins in alignment length of target protein2/runtime. Dashed lines indicate linear correlation, extrapolated from one thread. The whiskers extend to the most extreme data point that is less than 1.5-times the interquartile range from the box. The surprisingly clear correlation between the number of threads and speed demonstrates how well our implementation scales for multi-threading.
Similar articles
- COUSCOus: improved protein contact prediction using an empirical Bayes covariance estimator.
Rawi R, Mall R, Kunji K, El Anbari M, Aupetit M, Ullah E, Bensmail H. Rawi R, et al. BMC Bioinformatics. 2016 Dec 15;17(1):533. doi: 10.1186/s12859-016-1400-3. BMC Bioinformatics. 2016. PMID: 27978812 Free PMC article. - Improving accuracy of protein contact prediction using balanced network deconvolution.
Sun HP, Huang Y, Wang XF, Zhang Y, Shen HB. Sun HP, et al. Proteins. 2015 Mar;83(3):485-96. doi: 10.1002/prot.24744. Epub 2015 Jan 24. Proteins. 2015. PMID: 25524593 Free PMC article. - Improving residue-residue contact prediction via low-rank and sparse decomposition of residue correlation matrix.
Zhang H, Gao Y, Deng M, Wang C, Zhu J, Li SC, Zheng WM, Bu D. Zhang H, et al. Biochem Biophys Res Commun. 2016 Mar 25;472(1):217-22. doi: 10.1016/j.bbrc.2016.01.188. Epub 2016 Feb 23. Biochem Biophys Res Commun. 2016. PMID: 26920058 - MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins.
Jones DT, Singh T, Kosciolek T, Tetchner S. Jones DT, et al. Bioinformatics. 2015 Apr 1;31(7):999-1006. doi: 10.1093/bioinformatics/btu791. Epub 2014 Nov 26. Bioinformatics. 2015. PMID: 25431331 Free PMC article. - State-of-the-art bioinformatics protein structure prediction tools (Review).
Pavlopoulou A, Michalopoulos I. Pavlopoulou A, et al. Int J Mol Med. 2011 Sep;28(3):295-310. doi: 10.3892/ijmm.2011.705. Epub 2011 May 23. Int J Mol Med. 2011. PMID: 21617841 Review.
Cited by
- Deep-learning contact-map guided protein structure prediction in CASP13.
Zheng W, Li Y, Zhang C, Pearce R, Mortuza SM, Zhang Y. Zheng W, et al. Proteins. 2019 Dec;87(12):1149-1164. doi: 10.1002/prot.25792. Epub 2019 Aug 14. Proteins. 2019. PMID: 31365149 Free PMC article. - The EVcouplings Python framework for coevolutionary sequence analysis.
Hopf TA, Green AG, Schubert B, Mersmann S, Schärfe CPI, Ingraham JB, Toth-Petroczy A, Brock K, Riesselman AJ, Palmedo P, Kang C, Sheridan R, Draizen EJ, Dallago C, Sander C, Marks DS. Hopf TA, et al. Bioinformatics. 2019 May 1;35(9):1582-1584. doi: 10.1093/bioinformatics/bty862. Bioinformatics. 2019. PMID: 30304492 Free PMC article. - Inter-Residue Distance Prediction From Duet Deep Learning Models.
Zhang H, Huang Y, Bei Z, Ju Z, Meng J, Hao M, Zhang J, Zhang H, Xi W. Zhang H, et al. Front Genet. 2022 May 16;13:887491. doi: 10.3389/fgene.2022.887491. eCollection 2022. Front Genet. 2022. PMID: 35651930 Free PMC article. - Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13.
Hou J, Wu T, Cao R, Cheng J. Hou J, et al. Proteins. 2019 Dec;87(12):1165-1178. doi: 10.1002/prot.25697. Epub 2019 Apr 25. Proteins. 2019. PMID: 30985027 Free PMC article. - ConEVA: a toolbox for comprehensive assessment of protein contacts.
Adhikari B, Nowotny J, Bhattacharya D, Hou J, Cheng J. Adhikari B, et al. BMC Bioinformatics. 2016 Dec 7;17(1):517. doi: 10.1186/s12859-016-1404-z. BMC Bioinformatics. 2016. PMID: 27923350 Free PMC article.
References
- Rost B, Sander C. Bridging the protein sequence-structure gap by structure predictions. Annual review of biophysics and biomolecular structure. 1996;25:113–136. - PubMed
- Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjölander K, Ferrin TE, Burley SK, Sali A. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39(Database issue):D465–474. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources