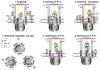Distinct docking and stabilization steps of the Pseudopilus conformational transition path suggest rotational assembly of type IV pilus-like fibers - PubMed (original) (raw)
Distinct docking and stabilization steps of the Pseudopilus conformational transition path suggest rotational assembly of type IV pilus-like fibers
Mangayarkarasi Nivaskumar et al. Structure. 2014.
Abstract
The closely related bacterial type II secretion (T2S) and type IV pilus (T4P) systems are sophisticated machines that assemble dynamic fibers promoting protein transport, motility, or adhesion. Despite their essential role in virulence, the molecular mechanisms underlying helical fiber assembly remain unknown. Here, we use electron microscopy and flexible modeling to study conformational changes of PulG pili assembled by the Klebsiella oxytoca T2SS. Neural network analysis of 3,900 pilus models suggested a transition path toward low-energy conformations driven by progressive increase in fiber helical twist. Detailed predictions of interprotomer contacts along this path were tested by site-directed mutagenesis, pilus assembly, and protein secretion analyses. We demonstrate that electrostatic interactions between adjacent protomers (P-P+1) in the membrane drive pseudopilin docking, while P-P+3 and P-P+4 contacts determine downstream fiber stabilization steps. These results support a model of a spool-like assembly mechanism for fibers of the T2SS-T4P superfamily.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no financial conflict of interest.
Figures
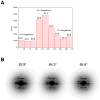
Figure 1. Variability of PulG filaments as seen by EM
(A) A reference-based approach was used to sort segments of PulG filaments extracted from EM images of negatively stained samples. Nine references were generated, having an axial rise of either 8.4, 10.4 or 12.4 Å and a twist of either 82.8°, 84.3° or 85.8°. Each reference was used to generate 36 different projections, involving increments of the azimuthal angle by 10° steps, for a total of 324 reference projections. The distribution found in (A) suggests that almost all of the variability is in the twist. The reality of this sorting can be tested by looking at averaged power spectra. (B) Averaged power spectra are shown for PulG segments selected from the central three bins of the distribution in (A). It can be seen that the near-equatorial layer line (Bessel order n=4) behaves exactly as expected for filaments with a rather fixed axial rise and a variable twist.
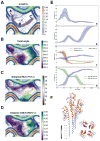
Figure 2. SOM analysis of 3901 PulG pilus models obtained from continuous distribution of angles between 81° and 88°
(A) U-matrix of the SOM clustering. The three main basins are labeled A, B and C. An ensemble of 100 possible transition paths is depicted in gray and red line indicates the corresponding mean path (as in B, C and D). (B) Projection of the twist-angle values (in degrees, purple/green color map) and the U-matrix (contour plot). (C)Projection of the distances between F1 and E5 in Å. (D) Projection of the distances between D48 and R87 in Å. (E) Evolution of the U-value (a), the twist-angle (b), relevant distances along the path between F1 and E5 (P-P+1 interface), K28 and E5 or K35 and E5 (P-P+3 interface) (c), and distances between D53 and K30 or K51 and D29 (P-P+4 interface) (d). Standard deviation is depicted as colored area around the mean curve of the 100 calculated transition paths shown in (A). (F) Side view (left) and top view (right) of PulG tetramer colored according to the rmsf per residue in Å, from red (rigid) to blue (flexible).
Figure 3. Representative PulG pilus structures in distinct transition path basins
Left, surface model of the PulG pilus with protomers P (orange), P+1 (green), P+3 (blue) and P+4 (maroon). Right, twist-angle evolution in the transition path showing top (above) and side views (below) of representative structures in basins A, B and C.
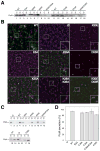
Figure 4. Functional analysis of P-P+1 interface
(A) Bacterial two-hybrid analysis graph showing mean values of β-galactosidase activity from 6 independent clones producing hybrid proteins with T18 and T25 CyaA fragments. (Ø), negative and (+) positive yeast leucine zipper control, WT, PulGWT or its variants, with residue positions and substitutions indicated. Error bars represent SD. Unpaired t test was used for statistical analysis: p<0.05 (**), p<0.005 (***), Ψ (no significant difference). (B) Schematic representation of T18- and T25-PulG hybrid orientation-dependent CyaA reconstitution leading to Lac+ (above) or Lac- phenotypes (below). (C) PulG immunodetection in 0.025 A600nm units of cells and pili fractions (C, S) of E. coli PAP7460 carrying pul genes on plasmid pCHAP8184 and pulG alleles as indicated. PulG* indicates a PulG adduct, probably resulting from oxidation. (D) PulA immunodetection in cell extracts and supernatants (C, S) of 0.005 A600nm units of E. coli PAP5299 carrying pul genes on plasmid pCHAP8184 and pulG alleles as indicated. (E) Cartoon PulG pilus model showing P−1-P interactions with E44-R88 and D48-R87 side chains shown as purple spheres.
Figure 5. Functional analysis of P-P+3 and P-P+4 interfaces
(A) PulG immunodetection in 0.005 A600nm of cell extract and sheared fractions (C, S) of E. coli expressing pul genes on plasmid pCHAP8184 and pulG alleles on pCHAP8658 and derivatives (Table S1). (B) Immunofluorescence microscopy of the E. coli strains as in (A). Cells stained with DAPI are shown in magenta and PulG pili, revealed with anti-PulG and Alexa-488 conjugated secondary antibodies, in green. Scale bar, 2 μm. (C) PulA immunodetection in cell extracts and supernatants (C, S) of 0.005 A600nm units of E. coli PAP5299 carrying pul genes on plasmid pCHAP8184 and pulG alleles as indicated. (D) Percentage of secreted PulA (mean + SD) from four independent experiments like in (C).
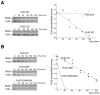
Figure 6. PulG turnover during protein secretion
(A) Left, strains PAP7232 (_pulG_WT) or PAP5327 (pulGE5A) were grown to late exponential phase in LB, 0.4% maltose. Protein synthesis was arrested by the addition of chloramphenicol (170 μg.ml−1) and 0.005 A600nm of bacterial extracts taken at indicated time points (min) were analyzed by SDS-PAGE and immuno-detection with anti-PulG and anti-RbsB antibodies. Right, levels of PulGWT (■) and PulGE5A (□) quantified using Image J, normalized against RbsB levels and plotted as a function of time. (B) Left, turnover PulGWT, PulGK30E and PulGK28A/K35A variants produced from low copy number plasmids in strain PAP7228 (ΔpulG). Protein synthesis was arrested by the addition of spectinomycin (100 μg.ml−1) and 0.005 A600nm of bacterial extracts taken at indicated time points (min) were analyzed by SDS-PAGE and immuno-detection with anti-PulG and anti-RbsB antibodies. Right, PulG levels normalized against RbsB levels, as a function of time. PulGWT (■), PulGK30E (◪) and PulGK28A/K35A (□).
Figure 7. The pseudopilus assembly model
Schematic representation of T2SS components labeled using the single letter code, with the secretin (D) in the outer membrane (OM), platform proteins F, C, L and M and assembly ATPase E in the inner membrane (IM). (1) Major pseudopilin G (in green) is targeted to the minor pseudopilins HIJK (in pink), and to assembly factors E and F. Variant GE5A (in grey) is defective in this step. (2) GspG protomer P (in blue) docks on the P+1 (in green) at the assembly site via electrostatic contacts (+ and − signs). (3) ATP hydrolysis promotes rotation of F in complex with nascent pseudopilus, spooling P into the fiber to add 10.4 Å to the polymer. (4) Top view showing ATPase E (grey circle with 6 segments) and F (crescent shape), surrounded by CLM complexes (small circles). Cycles of ATP binding, hydrolysis and release (red, yellow and white stars, respectively) drive rotation of F and assembly of G subunits at the base of the growing fiber. (5) After three cycles, P+3 is extracted from the membrane and stabilized through E5P+3 interactions with K28 and K35. (6) The next elongation step allows P-P+4 interactions to stabilize T2S pili in high twist (via K51P-E29P+4) or low twist (D53P - K30P+4) states.
Comment in
- A new twist in the assembly of type IV pilus-like fibers.
Burrows LL. Burrows LL. Structure. 2014 May 6;22(5):659-61. doi: 10.1016/j.str.2014.04.009. Structure. 2014. PMID: 24807073
Similar articles
- A new twist in the assembly of type IV pilus-like fibers.
Burrows LL. Burrows LL. Structure. 2014 May 6;22(5):659-61. doi: 10.1016/j.str.2014.04.009. Structure. 2014. PMID: 24807073 - Detailed structural and assembly model of the type II secretion pilus from sparse data.
Campos M, Nilges M, Cisneros DA, Francetic O. Campos M, et al. Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13081-6. doi: 10.1073/pnas.1001703107. Epub 2010 Jul 2. Proc Natl Acad Sci U S A. 2010. PMID: 20616068 Free PMC article. - Predicting Homogeneous Pilus Structure from Monomeric Data and Sparse Constraints.
Xiao K, Shu C, Yan Q, Sun X. Xiao K, et al. Biomed Res Int. 2015;2015:817134. doi: 10.1155/2015/817134. Epub 2015 May 4. Biomed Res Int. 2015. PMID: 26064954 Free PMC article. - Architecture of the type II secretion and type IV pilus machineries.
Ayers M, Howell PL, Burrows LL. Ayers M, et al. Future Microbiol. 2010 Aug;5(8):1203-18. doi: 10.2217/fmb.10.76. Future Microbiol. 2010. PMID: 20722599 Review. - The structure and mechanism of the bacterial type II secretion system.
Naskar S, Hohl M, Tassinari M, Low HH. Naskar S, et al. Mol Microbiol. 2021 Mar;115(3):412-424. doi: 10.1111/mmi.14664. Epub 2020 Dec 29. Mol Microbiol. 2021. PMID: 33283907 Review.
Cited by
- Identification of binding sites and favorable ligand binding moieties by virtual screening and self-organizing map analysis.
Harigua-Souiai E, Cortes-Ciriano I, Desdouits N, Malliavin TE, Guizani I, Nilges M, Blondel A, Bouvier G. Harigua-Souiai E, et al. BMC Bioinformatics. 2015 Mar 21;16:93. doi: 10.1186/s12859-015-0518-z. BMC Bioinformatics. 2015. PMID: 25888251 Free PMC article. - Direct interactions between the secreted effector and the T2SS components GspL and GspM reveal a new effector-sensing step during type 2 secretion.
Michel-Souzy S, Douzi B, Cadoret F, Raynaud C, Quinton L, Ball G, Voulhoux R. Michel-Souzy S, et al. J Biol Chem. 2018 Dec 14;293(50):19441-19450. doi: 10.1074/jbc.RA117.001127. Epub 2018 Oct 18. J Biol Chem. 2018. PMID: 30337370 Free PMC article. - Conserved Streptococcus pneumoniae spirosomes suggest a single type of transformation pilus in competence.
Laurenceau R, Krasteva PV, Diallo A, Ouarti S, Duchateau M, Malosse C, Chamot-Rooke J, Fronzes R. Laurenceau R, et al. PLoS Pathog. 2015 Apr 15;11(4):e1004835. doi: 10.1371/journal.ppat.1004835. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25876066 Free PMC article. - Dynamics of a type 2 secretion system pseudopilus unraveled by complementary approaches.
Bardiaux B, Cordier F, Brier S, López-Castilla A, Izadi-Pruneyre N, Nilges M. Bardiaux B, et al. J Biomol NMR. 2019 Jul;73(6-7):293-303. doi: 10.1007/s10858-019-00246-4. Epub 2019 May 23. J Biomol NMR. 2019. PMID: 31124002 Free PMC article. - Predicting and Interpreting the Structure of Type IV Pilus of Electricigens by Molecular Dynamics Simulations.
Shu C, Xiao K, Cao C, Ding D, Sun X. Shu C, et al. Molecules. 2017 Aug 12;22(8):1342. doi: 10.3390/molecules22081342. Molecules. 2017. PMID: 28805699 Free PMC article.
References
- Aas FE, Winter-Larsen HC, Wolfgang M, Frye S, Lovold C, Roos N, van Putten JPM, Koomey M. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect type IV pilus assembly, dynamics and associated functions. Mol. Microbiol. 2007;63:69–85. - PubMed
- Albers SV, Pohlschroder M. Diversity of archaeal type IV pilin-like structures. Extremophiles. 2009;13:403–410. - PubMed
- Ariga T, Muneyuki E, Yoshida M. F1-ATPase rotates by an asymmetric sequential mechanism using all three catalytic subunits. Nat Struct Mol Biol. 2007;14:841–846. - PubMed
- Ayers M, Howell PL, Burrows LL. Architecture of the type II secretion and type IV pilus machineries. Fut Microbiol. 2010;5:1203–1218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous