Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication - PubMed (original) (raw)
Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication
Wei-Chun Tang et al. J Virol. 2014 Jun.
Abstract
Positive-sense RNA viruses, such as dengue virus (DENV), hijack the intracellular membrane machinery for their own replication. The Rab18 protein, a member of the Rab GTPase family, key regulators of membrane trafficking, is located on the organelles involved in DENV infection, such as the endoplasmic reticulum (ER) and lipid droplets (LDs). In this study, we addressed the potential involvement of Rab18 in DENV infection by using cells overexpressing the wild-type, GTP-bound active form, or GDP-bound inactive form of Rab18 and cells with Rab18 knockdown. DENV replication, measured by viral protein, viral RNA, and viral progeny production, as well as LD induction, was reduced in cells with inactive Rab18 and in cells deprived of Rab18 expression, suggesting a positive role of Rab18 in the DENV life cycle. Interestingly, the interaction of fatty acid synthase (FASN), a key lipogenic enzyme in lipid biosynthesis, with DENV NS3 protein relied on the conversion of the GDP-bound to the GTP-bound form of Rab18. Furthermore, the targeting of FASN to sites participating in DENV infection, such as the ER and LDs, depends on functional Rab18. Thus, Rab18-mediated membrane trafficking of FASN and NS3 facilitates DENV replication, probably by ensuring a sufficient and coordinated lipid supply for membrane proliferation and arrangement.
Importance: Infection by dengue virus (DENV), an important mosquito-borne virus threatening ∼40% of the world's population, can cause mild dengue fever or severe dengue hemorrhagic fever and dengue shock syndrome. The pathogenesis mechanisms of DENV-related diseases are not clear, but high viral replication is believed to be a risk factor for the severe form of DENV infection. Thus, understanding the detailed mechanism of DENV replication might help address this devastating virus. Here, we found that Rab18, a small GTPase involved in vesicle trafficking and located in the endoplasmic reticulum network and on the surfaces of lipid droplets, positively regulates DENV replication. The functional machinery of Rab18 is required to recruit the enzyme fatty acid synthase to sites of DENV replication and to interact with DENV NS3 protein to promote fatty acid biosynthesis. Thus, DENV usurps Rab18 to facilitate its own replication.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
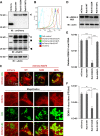
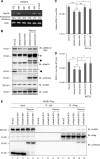
Reduced DENV-2 replication in cells with the GDP-bound inactive form of the Rab18 mutant. (A) Western blot (IB) analysis of Rab18 and mCherry proteins in human A549 cells expressing mCherry (vector control) or mCherry fused with the WT, GDP-bound mutant (S22N), or GTP-bound mutant (Q67L) of Rab18 with the indicated antibodies; actin was a loading control. The arrows indicate the positions of full-length mCherry-Rab18, the square indicates what is probably a degraded form of mCherry-Rab18 only seen in cells with mCherry-Rab18 overexpression, the arrowhead indicates the endogenous Rab18, and the asterisks mark background bands noted in all lanes. (B) Fluorescence (mCherry) expression of cells detected by flow cytometry. (C) Confocal microscopy of cells stained with Bodipy 493/503 to visualize LDs. Enlarged images of the boxed regions are shown below for better visualization of the locations of LDs and mCherry signal. (D to F) Cells were infected with DENV-2 (MOI = 5) for 24 h. (D) Western blot analysis of the DENV-2 NS3 protein level; actin was a loading control. (E) Quantitative RT-PCR analysis of the relative levels of DENV-2 RNA normalized to that of GAPDH (n = 3 per group). (F) Virus titration in culture supernatants by plaque-forming assay. The data are means and standard deviations (SD) (n = 3 per group). **, P < 0.01; ***, P < 0.001.
FIG 2
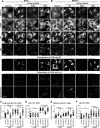
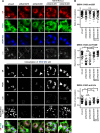
Rab18 regulates the intracellular distribution of FASN. (A and B) A549 cells with mCherry or mCherry fused with Rab18-WT, -S22N, or -Q67L were mock infected (A) or DENV-2 infected (MOI = 20 for 12 h) (B). The cells were immunofluorescently stained with anti-FASN plus Alexa Fluor 405 goat anti-mouse, anti-calreticulin (ER marker) plus Alexa Fluor 647 goat anti-chicken, and Bodipy 493/503. The bottom row shows magnifications of the boxed areas in the row above. (C and E) Colocalization coefficients of FASN with ER in the cells were quantified by use of ZEN 2011 software (Zeiss). (D and F) Quantification of LDs stained with Bodipy 493/503. Cells were cultured in medium with 10% FBS (A to D) or with 2% FBS (E and F). Means ± SD were calculated from 30 cells for each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
FIG 3
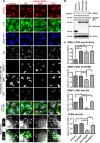
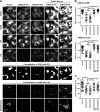
Rab18 activity is required for colocalization of DENV-2 NS3 with FASN and LDs. A549 cells with mCherry or mCherry fused with Rab18-WT, -S22N, or -Q67L were infected with DENV-2 (MOI = 20) for 12 h. (A) Cells were immunofluorescently stained with anti-FASN plus Alexa Fluor 405 goat anti-rabbit, anti-DENV-2 NS3 plus Alexa Fluor 647 goat anti-mouse, and Bodipy 493/503. Colocalization of the indicated proteins/organelle was visualized by use of ZEN 2011 software (Zeiss). The enlarged images of the boxed regions are shown for better visualization of DENV-2 NS3 wrapping around LDs (indicated by arrows). (B) Western blot analysis of FASN, DENV-2 NS3, mCherry, and actin. (C) Colocalization of DENV-2 NS3 with mCherry (Rab18), FASN, and LDs, as well as that of FASN with LDs, was measured by PCC. Means and SD were calculated from 30 cells for each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 4
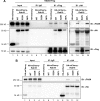
Rab18 is involved in the interaction of DENV-2 NS3 with FASN. HEK293T/17 cells were cotransfected with (A) or without (B) NS2B3-Flag plus the indicated plasmids expressing HA-mCherry or HA-mCherry-Rab18-WT, -S22N, or -Q67L for 48 h. The cell lysates were immunoprecipitated with control IgG, anti-Flag affinity gel, or anti-HA affinity gel. The immunoprecipitated proteins were then immunoblotted with antibodies against FASN, HA tag, and Flag tag as indicated.
FIG 5
Knockdown of Rab18 reduces DENV-2 replication and the interaction of NS3 with FASN. A549 cells were transduced with lentivirus expressing shRNA targeting Rab18 (shRab18) or shLacZ as a control. (A) Reverse transcription-PCR to verify the knockdown efficiency of shRab18 with primers specific for Rab18 and actin control. (B to D) The indicated cells were infected with DENV-2 (MOI = 5) for 24 h. (B) Western blot analysis of DENV-2 NS3, Rab18, mCherry, FASN, and actin. The positions of full-length and degraded mCherry-Rab18 (arrow and square), endogenous Rab18 (arrowhead), and background bands (asterisks) are marked. (C) Quantitative RT-PCR analysis of the relative levels of DENV-2 RNA normalized to that of GAPDH (n = 3 per group). (D) Virus titration in culture supernatants by plaque-forming assay. The data are means and SD (n = 3 per group). *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. (E) HEK293T/17 cells transfected with shLacZ or shRab18 (979 or 981) were selected by puromycin for 3 days and then were cotransfected with NS2B3-Flag plus mCherry or mCherry-Rab18-rescue for 24 h. The cell lysates were immunoprecipitated with control IgG or anti-Flag affinity gel. The immunoprecipitated proteins were immunoblotted with antibodies against FASN, Flag-tag, and mCherry as indicated.
FIG 6
Depletion of Rab18 decreases the colocalization of DENV-2 NS3 with FASN and LDs. (A) Cells were infected with DENV-2 (MOI = 20 for 12 h) and immunofluorescently stained with anti-calreticulin (ER marker) plus Alexa Fluor 594 goat anti-chicken, anti-FASN plus Alexa Fluor 405 goat anti-rabbit, anti-DENV-2 NS3 plus Alexa Fluor 647 goat anti-mouse, and Bodipy 493/503 (LD marker). (B) Quantification of DENV-2 NS3 colocalized with ER, FASN, and LDs analyzed with ZEN 2011 software (Zeiss). The data are means ± SD from 30 cells for each group. *, P < 0.05; ***, P < 0.001; n.s., not significant.
FIG 7
Rab18 knockdown restricts the redistribution of FASN to the ER and LDs in DENV-2-infected cells. (A) Cells were infected with DENV-2 (MOI = 20) for 12 h and immunofluorescently stained with anti-Rab18 plus Alexa Fluor 568 goat anti-rabbit, anti-FASN plus Alexa Fluor 405 goat anti-mouse, anti-calreticulin (ER marker) plus Alexa Fluor 647 goat anti-chicken, and Bodipy 493/503 (LD marker). Colocalization of FASN with the ER and LDs was analyzed with ZEN 2011 software (Zeiss). (B) Quantification of FASN colocalized with the ER and LDs was analyzed by ZEN 2011 software (Zeiss). (C) Quantification of LDs stained with Bodipy 493/503. The data are means ± SD from 30 cells for each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
Similar articles
- Fatty Acid Synthase Is Involved in Classical Swine Fever Virus Replication by Interaction with NS4B.
Liu YY, Liang XD, Liu CC, Cheng Y, Chen H, Baloch AS, Zhang J, Go YY, Zhou B. Liu YY, et al. J Virol. 2021 Aug 10;95(17):e0078121. doi: 10.1128/JVI.00781-21. Epub 2021 Aug 10. J Virol. 2021. PMID: 34132567 Free PMC article. - Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis.
Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Heaton NS, et al. Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17345-50. doi: 10.1073/pnas.1010811107. Epub 2010 Sep 20. Proc Natl Acad Sci U S A. 2010. PMID: 20855599 Free PMC article. - A Combined Genetic-Proteomic Approach Identifies Residues within Dengue Virus NS4B Critical for Interaction with NS3 and Viral Replication.
Chatel-Chaix L, Fischl W, Scaturro P, Cortese M, Kallis S, Bartenschlager M, Fischer B, Bartenschlager R. Chatel-Chaix L, et al. J Virol. 2015 Jul;89(14):7170-86. doi: 10.1128/JVI.00867-15. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926641 Free PMC article. - The Transactions of NS3 and NS5 in Flaviviral RNA Replication.
Tay MYF, Vasudevan SG. Tay MYF, et al. Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review. - The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity.
Lescar J, Soh S, Lee LT, Vasudevan SG, Kang C, Lim SP. Lescar J, et al. Adv Exp Med Biol. 2018;1062:115-129. doi: 10.1007/978-981-10-8727-1_9. Adv Exp Med Biol. 2018. PMID: 29845529 Review.
Cited by
- Diacylglycerol O-acyltransferase 2, a Novel Target of Flavivirus NS2B3 Protease, Promotes Zika Virus Replication by Regulating Lipid Droplet Formation.
Luo X, Yuan Y, Ma X, Luo X, Chen J, Chen C, Yang X, Yang J, Zhu X, Li M, Liu Y, Zhang P, Liu C. Luo X, et al. Research (Wash D C). 2024 Oct 24;7:0511. doi: 10.34133/research.0511. eCollection 2024. Research (Wash D C). 2024. PMID: 39449854 Free PMC article. - The triglyceride-synthesizing enzyme diacylglycerol acyltransferase 2 modulates the formation of the hepatitis C virus replication organelle.
Reichert I, Lee JY, Weber L, Fuh MM, Schlaeger L, Rößler S, Kinast V, Schlienkamp S, Conradi J, Vondran FWR, Pfaender S, Scaturro P, Steinmann E, Bartenschlager R, Pietschmann T, Heeren J, Lauber C, Vieyres G. Reichert I, et al. PLoS Pathog. 2024 Sep 6;20(9):e1012509. doi: 10.1371/journal.ppat.1012509. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39241103 Free PMC article. - Roles of lipid droplets and related proteins in metabolic diseases.
Zhang Z, Yu Z, Liang D, Song K, Kong X, He M, Liao X, Huang Z, Kang A, Bai R, Ren Y. Zhang Z, et al. Lipids Health Dis. 2024 Jul 19;23(1):218. doi: 10.1186/s12944-024-02212-y. Lipids Health Dis. 2024. PMID: 39030618 Free PMC article. Review. - Dengue virus non-structural protein 3 inhibits mitochondrial respiration by impairing complex I function.
Sousa BG, Mebus-Antunes NC, Fernandes-Siqueira LO, Caruso MB, Saraiva GN, Carvalho CF, Neves-Martins TC, Galina A, Zingali RB, Zeidler JD, Da Poian AT. Sousa BG, et al. mSphere. 2024 Jul 30;9(7):e0040624. doi: 10.1128/msphere.00406-24. Epub 2024 Jul 9. mSphere. 2024. PMID: 38980068 Free PMC article. - Pseudorabies virus hijacks the Rab6 protein to promote viral assembly and egress.
Liang DG, Guo YK, Zhao SB, Yang GY, Han YQ, Chu BB, Ming SL. Liang DG, et al. Vet Res. 2024 May 28;55(1):68. doi: 10.1186/s13567-024-01328-4. Vet Res. 2024. PMID: 38807225 Free PMC article.
References
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous