Correlative light- and electron microscopy with chemical tags - PubMed (original) (raw)
. 2014 May;186(2):205-13.
doi: 10.1016/j.jsb.2014.03.018. Epub 2014 Mar 31.
Michael Kunz 1, Ulrike Endesfelder 2, Stefanie Bunse 3, Christoph Wigge 1, Zhou Yu 1, Victor-Valentin Hodirnau 1, Margot P Scheffer 1, Anja Seybert 1, Sebastian Malkusch 2, Erin M Schuman 3, Mike Heilemann 2, Achilleas S Frangakis 4
Affiliations
- PMID: 24698954
- PMCID: PMC5321482
- DOI: 10.1016/j.jsb.2014.03.018
Correlative light- and electron microscopy with chemical tags
Mario Perkovic et al. J Struct Biol. 2014 May.
Abstract
Correlative microscopy incorporates the specificity of fluorescent protein labeling into high-resolution electron micrographs. Several approaches exist for correlative microscopy, most of which have used the green fluorescent protein (GFP) as the label for light microscopy. Here we use chemical tagging and synthetic fluorophores instead, in order to achieve protein-specific labeling, and to perform multicolor imaging. We show that synthetic fluorophores preserve their post-embedding fluorescence in the presence of uranyl acetate. Post-embedding fluorescence is of such quality that the specimen can be prepared with identical protocols for scanning electron microscopy (SEM) and transmission electron microscopy (TEM); this is particularly valuable when singular or otherwise difficult samples are examined. We show that synthetic fluorophores give bright, well-resolved signals in super-resolution light microscopy, enabling us to superimpose light microscopic images with a precision of up to 25 nm in the x-y plane on electron micrographs. To exemplify the preservation quality of our new method we visualize the molecular arrangement of cadherins in adherens junctions of mouse epithelial cells.
Keywords: Adherens junctions; Correlative electron and light microscopy; Electron tomography.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Fig.1
Live-cell imaging and post-embedding fluorescence at different UA levels. (a, b) Live-cell imaging of L-cells stably expressing SNAPf-NCadherin labeled with Alexa Fluor 647. The cell–cell junctions are visualized in the red channel. The auto-fluorescence is shown in the green channel. (c, d) The corresponding fluorescence within the resin-block after high-pressure freezing and freeze-substitution with 0.1% and 4% UA. (e) Fluorescence intensity measured along the cell junctions in live-cell imaging (dark red), postembedding in the resin-block (light red), auto-fluorescence measured in the green channel in live-cell imaging (dark green), and the auto-fluorescence postembedding in the resin block (light green). At least three regions per uranyl acetate concentration were evaluated with 200 sample points each. Error bars represent the standard deviation of the intensity along the junction. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig.2
ia-SEM CLEM of NCadherin in L-cells. (a) Fluorescence image of L-cells stably expressing SNAPf-NCadherin labeled with Alexa Fluor 647 after high-pressure freezing and freeze substitution with 2% UA. The box outlines the 32 μm × 17 μm area enlarged in (b) and shows the interface of six neighboring cells. (c) Overlay of the confocal fluorescence image of SNAPf-NCadherin from (b) on a tomographic slice from the ia-SEM image stack showing the six neighboring cells outlined with different colors and numbered. The specific fluorescence of Alexa Fluor 647 shown in red is localized mainly at cell–cell contacts, and occasionally on the cell surface at densely clustered filopodia. The contrast in the SEM images allows the identification of sub-nuclear compartments: Nucleolus (Nu; medium grey), chromatin (Ch; light grey) and interchromatin compartment (IC; dark grey) as well as diverse cytoplasmic vesicles (V) and filopodia (F). (d) Volume representation of the same SEM dataset (32 μm × 17 μm × 10 μm), overlaid with the confocal fluorescence signal. The fluorescence is visualized by volume rendering in an orange heat color. The different cells are represented as transparent isosurfaces in corresponding colors to (c); blue (cell 1), purple (cell 2), grey-green (cell 3 and 6), and grey-blue (cell 4) with opaque nuclei shown in cells 1 and 2, corresponding to the contours in (c). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
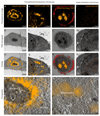
Fig.3
CLEM of various intracellular and intranuclear proteins. (a) FM image of SNAPf-H2B-TMR. (e) FM image of SNAPf-Cox8A-TMR. (i) Dual color FM of SNAPf-ADRbeta2-AF647 (red) and Halo-Nucleolin-TMR (orange). (l) Super-resolution microscopy of SNAPf-H2B-SiR (orange). (b, f, j, m) TEM micrographs after FM imaging. (c, g, k, n) overlay of the FM images with the corresponding TEM micrographs. (c) FM signal overlaying with the nucleus sparing the darker stained nucleolus. (g) FM signal overlaying with intracellular mitochondria. (k) FM signal overlaying with cellular filipodia (red) as well as with nucleoli (orange). (n) Super-resolution signal overlaying with the nucleus. (d) tomogram acquired on the boxed area in (c). FM signal (orange) is sparing the dark nucleolus. (h) tomogram acquired on the boxed area in (g). FM signal (orange) is localized on the mitochondria. Mitochondrial cristae as well as cytoskeletal elements and parts of the Golgi apparatus are visible. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig.4
Localization-based microscopy combined with ET, and sub-tomogram averaging of NCadherin expressing L-cells. (a) Superimposed images of the super-resolution signal of SNAPf-NCadherin (orange) and the corresponding TEM micrograph. Lower insert is showing the precision of the protein localization with super-resolution microscopy. The blue line depicts the measured profile across the cellular junction in the super-resolution image, whereas the red line is the Gaussian fit to it. The full width at half maximum of the Gaussian is 42 nm, which is in good agreement with calculated precision of the localization precision of the super-resolution microscopy with sigma equal to 17 nm (2.35 * sigma). Upper insert is showing the precision of proteins localization within the tomograms, on the example of the cadherin molecules. The blue plot shows the normalized intensity profile across the junction measured on the EM images, which shows a membrane spacing of ~26 nm (blue double arrow). The red plot shows the normalized intensity profile across the super-resolution signal. The shift between the red and the blue curve indicates the precision of protein localization within the EM tomogram. The shift (black double arrow) is measured between the peak of the super-resolution signal (red) and the center of the neighboring cell membranes (blue). The precision of the protein localization within the EM tomogram was estimated at ~25 nm. (b) Overlay of a 2 nm thick tomographic slice with the super-resolution signal (orange). The first fluorescent cluster is localized between two cell membranes with a spacing of ~26 nm. The second fluorescent cluster can be associated with the cell membrane of the left cell ~80 nm higher than the first and shows a gathering of cadherins, without a connection to the juxtaposed cell (Box 2). (c) Volume representation of intracellular organelles recognizable in the tomogram; mitochondria (yellow), vesicles (light blue), microtubules (green), cytoplasmic compounds (violet). (d) Sub-tomogram averaging of the first fluorescence cluster reveals two cell membranes at a distance of ~26 nm together with two densities spanning the distance between the membranes (white arrowheads). Volume representation of the membrane (light blue) and the obliquely oriented densities (yellow) with a thickness of 3 nm (black arrowheads). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Similar articles
- Correlative stochastic optical reconstruction microscopy and electron microscopy.
Kim D, Deerinck TJ, Sigal YM, Babcock HP, Ellisman MH, Zhuang X. Kim D, et al. PLoS One. 2015 Apr 15;10(4):e0124581. doi: 10.1371/journal.pone.0124581. eCollection 2015. PLoS One. 2015. PMID: 25874453 Free PMC article. - High-resolution imaging by scanning electron microscopy of semithin sections in correlation with light microscopy.
Koga D, Kusumi S, Shodo R, Dan Y, Ushiki T. Koga D, et al. Microscopy (Oxf). 2015 Dec;64(6):387-94. doi: 10.1093/jmicro/dfv042. Epub 2015 Jul 22. Microscopy (Oxf). 2015. PMID: 26206941 - FIB-SEM imaging properties of Drosophila melanogaster tissues embedded in Lowicryl HM20.
Porrati F, Grewe D, Seybert A, Frangakis AS, Eltsov M. Porrati F, et al. J Microsc. 2019 Feb;273(2):91-104. doi: 10.1111/jmi.12764. Epub 2018 Nov 12. J Microsc. 2019. PMID: 30417390 - Correlative video-light-electron microscopy: development, impact and perspectives.
Rizzo R, Parashuraman S, Luini A. Rizzo R, et al. Histochem Cell Biol. 2014 Aug;142(2):133-8. doi: 10.1007/s00418-014-1249-3. Epub 2014 Jul 17. Histochem Cell Biol. 2014. PMID: 25030356 Review. - Correlative microscopy.
Loussert Fonta C, Humbel BM. Loussert Fonta C, et al. Arch Biochem Biophys. 2015 Sep 1;581:98-110. doi: 10.1016/j.abb.2015.05.017. Epub 2015 Jun 10. Arch Biochem Biophys. 2015. PMID: 26072116 Review.
Cited by
- Assessing Intra-Bundle Impregnation in Partially Impregnated Glass Fiber-Reinforced Polypropylene Composites Using a 2D Extended-Field and Multimodal Imaging Approach.
Sidlipura S, Ayadi A, Lagardère Deléglise M. Sidlipura S, et al. Polymers (Basel). 2024 Jul 30;16(15):2171. doi: 10.3390/polym16152171. Polymers (Basel). 2024. PMID: 39125198 Free PMC article. - An aldehyde-crosslinking mitochondrial probe for STED imaging in fixed cells.
Chen J, Stephan T, Gaedke F, Liu T, Li Y, Schauss A, Chen P, Wulff V, Jakobs S, Jüngst C, Chen Z. Chen J, et al. Proc Natl Acad Sci U S A. 2024 May 7;121(19):e2317703121. doi: 10.1073/pnas.2317703121. Epub 2024 Apr 30. Proc Natl Acad Sci U S A. 2024. PMID: 38687792 Free PMC article. - Rapid in-EPON CLEM: Combining fast and efficient labeling of self-labeling enzyme tags with EM-resistant Janelia Fluor dyes and StayGold.
Franzkoch R, Wilkening S, Liss V, Holtmannspötter M, Kurre R, Psathaki OE, Hensel M. Franzkoch R, et al. Heliyon. 2024 Mar 18;10(7):e28055. doi: 10.1016/j.heliyon.2024.e28055. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38560224 Free PMC article. - Indirect Correlative Light and Electron Microscopy (iCLEM): A Novel Pipeline for Multiscale Quantification of Structure From Molecules to Organs.
Struckman HL, Moise N, Vanslembrouck B, Rothacker N, Chen Z, van Hengel J, Weinberg SH, Veeraraghavan R. Struckman HL, et al. Microsc Microanal. 2024 Apr 29;30(2):318-333. doi: 10.1093/mam/ozae021. Microsc Microanal. 2024. PMID: 38525890 - Correlative SIP-FISH-Raman-SEM-NanoSIMS links identity, morphology, biochemistry, and physiology of environmental microbes.
Schaible GA, Kohtz AJ, Cliff J, Hatzenpichler R. Schaible GA, et al. ISME Commun. 2022 Jun 30;2(1):52. doi: 10.1038/s43705-022-00134-3. ISME Commun. 2022. PMID: 37938730 Free PMC article.
References
- Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. - PubMed
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. - PubMed
- Canny J. A computational approach to edge-detection. IEEE Trans Pattern Anal Mach Intell. 1986;8:679–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources