Joint analysis of functional genomic data and genome-wide association studies of 18 human traits - PubMed (original) (raw)
Joint analysis of functional genomic data and genome-wide association studies of 18 human traits
Joseph K Pickrell. Am J Hum Genet. 2014.
Erratum in
- Am J Hum Genet. 2014 Jul 3;95(1):126
Abstract
Annotations of gene structures and regulatory elements can inform genome-wide association studies (GWASs). However, choosing the relevant annotations for interpreting an association study of a given trait remains challenging. I describe a statistical model that uses association statistics computed across the genome to identify classes of genomic elements that are enriched with or depleted of loci influencing a trait. The model naturally incorporates multiple types of annotations. I applied the model to GWASs of 18 human traits, including red blood cell traits, platelet traits, glucose levels, lipid levels, height, body mass index, and Crohn disease. For each trait, I used the model to evaluate the relevance of 450 different genomic annotations, including protein-coding genes, enhancers, and DNase-I hypersensitive sites in over 100 tissues and cell lines. The fraction of phenotype-associated SNPs influencing protein sequence ranged from around 2% (for platelet volume) up to around 20% (for low-density lipoprotein cholesterol), repressed chromatin was significantly depleted for SNPs associated with several traits, and cell-type-specific DNase-I hypersensitive sites were enriched with SNPs associated with several traits (for example, the spleen in platelet volume). Finally, reweighting each GWAS by using information from functional genomics increased the number of loci with high-confidence associations by around 5%.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
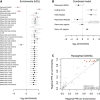
Application of the Model to HDL Cholesterol (A) Single-annotation models. I fit the model to each annotation individually, including a SNP-level effect for SNPs 0–5 kb from a TSS, a SNP-level effect for SNPs 5–10 kb from a TSS, a region-level effect for regions in the top third of gene density, and a region-level effect for regions in the bottom third of gene density. Plotted are the maximum-likelihood estimates and 95% CIs of the enrichment parameter for each annotation. Annotations are ordered according to how much they improved the likelihood of the model (at the top are those that improved the likelihood the most). In red are the annotations included in the joint model, and in pink are the annotations that are statistically equivalent to those included in the combined model. (B) Joint model. Using the algorithm described in the Material and Methods, I built a model combining multiple annotations. Shown are the maximum-likelihood estimates and 95% CIs of the enrichment effects of each annotation. Note that although these are the maximum-likelihood estimates, model choice was performed with a penalized likelihood. In parentheses next to each annotation (except for those relating to distance to TSSs) is the total number of annotations statistically equivalent to the included annotation in a conditional analysis. (C) Reweighted GWASs. I reweighted the GWASs by using the model with all the annotations in (B) (under the penalized enrichment parameters from Table S9). Each point represents a region of the genome, and shown are the posterior probabilities of association (PPAs) of the regions in the models with and without the annotations.
Figure 2
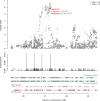
Regional Plot Surrounding NR0B2 The top panel shows a plot of the p values for association with HDL levels at each SNP in this region. In the middle panel is the fitted empirical prior probability (conditional on there being a single causal SNP in the region) that each SNP is the causal one in the region. This prior was estimated with the combined model with the annotations in Figure 1B. In the lower panel are the positions of the annotations included in the model. The reported p value indicated by an asterisk is for rs12748152, which has _r_2 = 0.85 with rs6659176.
Figure 3
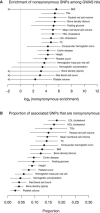
Estimated Role of Protein-Coding Changes in Each Trait (A) Estimated enrichment of nonsynonymous SNPs. For each trait, I fit a model including an effect of nonsynonymous SNPs and an effect of SNPs within 5 kb of a TSS. Shown are the estimated enrichment parameters and 95% CIs for the nonsynonymous SNPs. (B) Estimated proportion of GWAS hits driven by nonsynonymous SNPs. For each trait, using the model fit in (A), I estimated the proportion of GWAS signals driven by nonsynonymous SNPs. This estimate and its SE are shown.
Figure 4
Combined Models for Nine Traits For each trait, I built a combined model of annotations by using the algorithm presented in the Material and Methods. Shown are the maximum-likelihood estimates and 95% CIs for all annotations included in each model. Note that although these are the maximum-likelihood estimates, model choice was done with a penalized likelihood (Material and Methods). For the other nine traits, see Figure S14. In parentheses next to each annotation (except for those relating to distance to TSSs) is the total number of annotations that are statistically equivalent to the included annotation in a conditional analysis (Material and Methods). The annotation of DNase-I hypersensitive sites in fetal fibroblasts from the abdomen (marked by an asterisk) had a positive effect when treated alone; see the main text for discussion.
Similar articles
- Comprehensive evaluation of disease- and trait-specific enrichment for eight functional elements among GWAS-identified variants.
Markunas CA, Johnson EO, Hancock DB. Markunas CA, et al. Hum Genet. 2017 Jul;136(7):911-919. doi: 10.1007/s00439-017-1815-6. Epub 2017 May 31. Hum Genet. 2017. PMID: 28567521 - Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation.
Croteau-Chonka DC, Rogers AJ, Raj T, McGeachie MJ, Qiu W, Ziniti JP, Stubbs BJ, Liang L, Martinez FD, Strunk RC, Lemanske RF Jr, Liu AH, Stranger BE, Carey VJ, Raby BA. Croteau-Chonka DC, et al. PLoS One. 2015 Oct 16;10(10):e0140758. doi: 10.1371/journal.pone.0140758. eCollection 2015. PLoS One. 2015. PMID: 26474488 Free PMC article. Review. - Comprehensive identification of pleiotropic loci for body fat distribution using the NHGRI-EBI Catalog of published genome-wide association studies.
Kaur Y, Wang DX, Liu HY, Meyre D. Kaur Y, et al. Obes Rev. 2019 Mar;20(3):385-406. doi: 10.1111/obr.12806. Epub 2018 Nov 22. Obes Rev. 2019. PMID: 30565845 Review. - SNP eQTL status and eQTL density in the adjacent region of the SNP are associated with its statistical significance in GWA studies.
Gorlov I, Xiao X, Mayes M, Gorlova O, Amos C. Gorlov I, et al. BMC Genet. 2019 Nov 12;20(1):85. doi: 10.1186/s12863-019-0786-0. BMC Genet. 2019. PMID: 31718536 Free PMC article. - An efficient unified model for genome-wide association studies and genomic selection.
Li H, Su G, Jiang L, Bao Z. Li H, et al. Genet Sel Evol. 2017 Aug 24;49(1):64. doi: 10.1186/s12711-017-0338-x. Genet Sel Evol. 2017. PMID: 28836943 Free PMC article.
Cited by
- Predicting cell type-specific epigenomic profiles accounting for distal genetic effects.
Murphy AE, Beardall W, Rei M, Phuycharoen M, Skene NG. Murphy AE, et al. Nat Commun. 2024 Nov 16;15(1):9951. doi: 10.1038/s41467-024-54441-5. Nat Commun. 2024. PMID: 39550354 Free PMC article. - Decoding mutational hotspots in human disease through the gene modules governing thymic regulatory T cells.
Raposo AASF, Rosmaninho P, Silva SL, Paço S, Brazão ME, Godinho-Santos A, Tokunaga-Mizoro Y, Nunes-Cabaço H, Serra-Caetano A, Almeida ARM, Sousa AE. Raposo AASF, et al. Front Immunol. 2024 Oct 15;15:1458581. doi: 10.3389/fimmu.2024.1458581. eCollection 2024. Front Immunol. 2024. PMID: 39483472 Free PMC article. - First insight of the genome-wide association study and genomic prediction into enteritis disease (Vibrio harveyi) resistance trait in the lined seahorse (Hippocampus erectus).
Li S, Liu X, Shen F, Lin T, Zhang D. Li S, et al. Front Immunol. 2024 Oct 3;15:1474746. doi: 10.3389/fimmu.2024.1474746. eCollection 2024. Front Immunol. 2024. PMID: 39421751 Free PMC article. - Transcripts with high distal heritability mediate genetic effects on complex metabolic traits.
Tyler AL, Mahoney JM, Keller MP, Baker CN, Gaca M, Srivastava A, Gerdes Gyuricza I, Braun MJ, Rosenthal NA, Attie AD, Churchill GA, Carter GW. Tyler AL, et al. bioRxiv [Preprint]. 2024 Sep 27:2024.09.26.613931. doi: 10.1101/2024.09.26.613931. bioRxiv. 2024. PMID: 39386475 Free PMC article. Preprint. - A Deep Dive into Statistical Modeling of RNA Splicing QTLs Reveals New Variants that Explain Neurodegenerative Disease.
Wang D, Gazzara MR, Jewell S, Wales-McGrath B, Brown CD, Choi PS, Barash Y. Wang D, et al. bioRxiv [Preprint]. 2024 Sep 3:2024.09.01.610696. doi: 10.1101/2024.09.01.610696. bioRxiv. 2024. PMID: 39282456 Free PMC article. Preprint.
References
- Schork A.J., Thompson W.K., Pham P., Torkamani A., Roddey J.C., Sullivan P.F., Kelsoe J.R., O’Donovan M.C., Furberg H., Schork N.J., Tobacco and Genetics Consortium. Bipolar Disorder Psychiatric Genomics Consortium. Schizophrenia Psychiatric Genomics Consortium All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources