Non-specificity of Pitstop 2 in clathrin-mediated endocytosis - PubMed (original) (raw)
Non-specificity of Pitstop 2 in clathrin-mediated endocytosis
Anna K Willox et al. Biol Open. 2014.
Abstract
Small molecule inhibitors of clathrin-mediated endocytosis are highly desired for the dissection of membrane trafficking pathways in the lab and for potential use as anti-infectives in the clinic. One inhibition strategy is to prevent clathrin from contacting adaptor proteins so that clathrin-mediated endocytosis cannot occur. "Pitstop" compounds have been developed that block only one of the four functional interaction sites on the N-terminal domain of clathrin heavy chain. Despite this limitation, Pitstop 2 causes profound inhibition of clathrin-mediated endocytosis. In this study, we probed for non-specific activity of Pitstop 2 by examining its action in cells expressing clathrin heavy chain harbouring mutations in the N-terminal domain interaction sites. We conclude that the inhibition observed with this compound is due to non-specificity, i.e. it causes inhibition away from its proposed mode of action. We recommend that these compounds be used with caution in cells and that they should not be used to conclude anything of the function of clathrin's N-terminal domain.
Keywords: Clathrin; Endocytosis; Pitstop; Small-molecule inhibitor.
© 2014. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests: The authors have no competing interests to declare.
Figures
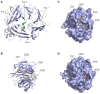
Fig. 1.. Structural view of interactions at the N-terminal domain of clathrin.
(A) Structural model of CHC(1–363), comprising the seven-bladed β-propeller (blue, 1–330) and linker (grey). The interaction sites listed in Table 1 are shown together with their respective ligands. The model is an alignment of three separate structures 1UTC, 1C9I and 3GD1. The blades of the β-propeller are numbered (β1–β7). (B) Cartoon view of the clathrin-box motif interaction site (Site 1). The ligand AVSLLDLA from β3-subunit of AP-3 (PDB code 1C9I) is shown together with residues that were targeted for the C+ mutation (Table 1). (C) Surface view of the N-terminal domain in the same orientation as panel B, showing the clathrin-box motif peptide binding in the groove between blades 1 and 2. Positions of the residues targeted for mutation C+ are indicated. (D) Same view as panel C, but showing the NTD in complex with Pitstop 2 (PDB code 4G55). Pitstop 2 occupies the groove that clathrin-box motif-containing peptides bind, and this is believed to be its mode of action. No significant density for the ligand is found elsewhere on the NTD (von Kleist et al., 2011). Note that in 4G55, two alternative positions for side chains of Lys96 and Lys98 are present, conformation B is shown.
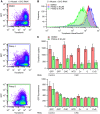
Fig. 2.. Pitstop 2 inhibits clathrin-mediated endocytosis non-specifically.
(A) Typical data from transferrin uptake experiments with clathrin-depleted cells expressing GFP-CHC(1–1675) (C+ mutant). Cells were treated with DMSO or Pitstop 1 (30 µM) or Pitstop 2 (30 µM) for 30 min during serum starvation, as indicated. A pseudocoloured bivariate histogram is shown for GFP intensity versus transferrin-Alexa647 fluorescence. The GFP-positive cells were a clearly discerned population that could be gated and analysed as indicated. (B) Histogram to show the frequency of clathrin-depleted cells with a given transferrin uptake expressing full-length RNAi-resistant GFP-tagged CHC harbouring the C+ mutations. Histograms were generated from data gated as indicated in panel A. Note the logarithmic scale on the _x_-axis. (C) Bar charts to summarise transferrin uptake experiments. Cells were transfected as indicated. Median transferrin-Alexa647 (above) or GFP (below) fluorescence values from cells gated for GFP expression are shown as mean ± s.e.m. from four independent experiments.
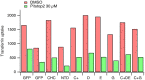
Fig. 3.. The inhibition of CME by Pitstop 2 is not confined to the clathrin-box site.
Bar chart of a single transferrin uptake experiment. Cells were transfected as indicated. Median transferrin-Alexa647 fluorescence values from cells gated for GFP expression are shown.
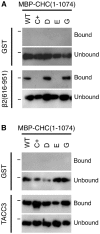
Fig. 4.. Mutation of the clathrin-box site is sufficient to block interaction with clathrin-box motif-containing proteins.
Results of a typical binding experiment to analyse the role of the four interaction sites on the NTD of CHC. Wild-type MBP-CHC(1–1074)-His6 or indicated mutants were incubated with (A) GST or GST-β2 adaptin (616–951) or (B) GST or GST-TACC3-His6 that were phosphorylated with Aurora A/TPX2, before pull-down on glutathione beads. Bound, and a sample of unbound, material was separated by SDS-PAGE and transferred to nitrocellulose for western blotting using an anti-CHC (TD.1) antibody. Markers indicate position of 150 kDa.
Similar articles
- Inhibition Clathrin Mediated Endocytosis: Pitstop 1 and Pitstop 2 Chimeras.
Prichard K, Chau N, Xue J, Krauss M, Sakoff JA, Gilbert J, Bahnik C, Muehlbauer M, Radetzki S, Robinson PJ, Haucke V, McCluskey A. Prichard K, et al. ChemMedChem. 2024 Oct 16;19(20):e202400253. doi: 10.1002/cmdc.202400253. Epub 2024 Sep 29. ChemMedChem. 2024. PMID: 38894585 - Pitstop 2 is a potent inhibitor of clathrin-independent endocytosis.
Dutta D, Williamson CD, Cole NB, Donaldson JG. Dutta D, et al. PLoS One. 2012;7(9):e45799. doi: 10.1371/journal.pone.0045799. Epub 2012 Sep 21. PLoS One. 2012. PMID: 23029248 Free PMC article. - Clathrin-mediated integrin αIIbβ3 trafficking controls platelet spreading.
Gao W, Shi P, Chen X, Zhang L, Liu J, Fan X, Luo X. Gao W, et al. Platelets. 2018 Sep;29(6):610-621. doi: 10.1080/09537104.2017.1353682. Epub 2017 Sep 29. Platelets. 2018. PMID: 28961039 - Getting in touch with the clathrin terminal domain.
Lemmon SK, Traub LM. Lemmon SK, et al. Traffic. 2012 Apr;13(4):511-9. doi: 10.1111/j.1600-0854.2011.01321.x. Epub 2012 Jan 13. Traffic. 2012. PMID: 22239657 Free PMC article. Review. - Clathrin's life beyond 40: Connecting biochemistry with physiology and disease.
Briant K, Redlingshöfer L, Brodsky FM. Briant K, et al. Curr Opin Cell Biol. 2020 Aug;65:141-149. doi: 10.1016/j.ceb.2020.06.004. Epub 2020 Aug 21. Curr Opin Cell Biol. 2020. PMID: 32836101 Review.
Cited by
- Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification.
Dejonghe W, Kuenen S, Mylle E, Vasileva M, Keech O, Viotti C, Swerts J, Fendrych M, Ortiz-Morea FA, Mishev K, Delang S, Scholl S, Zarza X, Heilmann M, Kourelis J, Kasprowicz J, Nguyen le SL, Drozdzecki A, Van Houtte I, Szatmári AM, Majda M, Baisa G, Bednarek SY, Robert S, Audenaert D, Testerink C, Munnik T, Van Damme D, Heilmann I, Schumacher K, Winne J, Friml J, Verstreken P, Russinova E. Dejonghe W, et al. Nat Commun. 2016 Jun 8;7:11710. doi: 10.1038/ncomms11710. Nat Commun. 2016. PMID: 27271794 Free PMC article. - Sterolight as imaging tool to study sterol uptake, trafficking and efflux in living cells.
Králová J, Popr M, Valečka J, Bartůněk P. Králová J, et al. Sci Rep. 2022 Apr 15;12(1):6264. doi: 10.1038/s41598-022-10134-x. Sci Rep. 2022. PMID: 35428843 Free PMC article. - Initial ciliary assembly in Chlamydomonas requires Arp2/3 complex-dependent endocytosis.
Bigge BM, Rosenthal NE, Avasthi P. Bigge BM, et al. Mol Biol Cell. 2023 Apr 1;34(4):ar24. doi: 10.1091/mbc.E22-09-0443. Epub 2023 Feb 8. Mol Biol Cell. 2023. PMID: 36753382 Free PMC article. - Diffusional spread and confinement of newly exocytosed synaptic vesicle proteins.
Gimber N, Tadeus G, Maritzen T, Schmoranzer J, Haucke V. Gimber N, et al. Nat Commun. 2015 Sep 24;6:8392. doi: 10.1038/ncomms9392. Nat Commun. 2015. PMID: 26399746 Free PMC article. - Role of Clathrin and Dynamin in Clathrin Mediated Endocytosis/Synaptic Vesicle Recycling and Implications in Neurological Diseases.
Prichard KL, O'Brien NS, Murcia SR, Baker JR, McCluskey A. Prichard KL, et al. Front Cell Neurosci. 2022 Jan 18;15:754110. doi: 10.3389/fncel.2021.754110. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35115907 Free PMC article. Review.
References
- Collette J. R., Chi R. J., Boettner D. R., Fernandez-Golbano I. M., Plemel R., Merz A. J., Geli M. I., Traub L. M., Lemmon S. K. (2009). Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs. Mol. Biol. Cell 20, 3401–3413 10.1091/mbc.E08-10-1082 - DOI - PMC - PubMed
- Edeling M. A., Mishra S. K., Keyel P. A., Steinhauser A. L., Collins B. M., Roth R., Heuser J. E., Owen D. J., Traub L. M. (2006). Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell 10, 329–342 10.1016/j.devcel.2006.01.016 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous