Exploiting polypharmacology for drug target deconvolution - PubMed (original) (raw)
Exploiting polypharmacology for drug target deconvolution
Taranjit Singh Gujral et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Polypharmacology (action of drugs against multiple targets) represents a tempting avenue for new drug development; unfortunately, methods capable of exploiting the known polypharmacology of drugs for target deconvolution are lacking. Here, we present an ensemble approach using elastic net regularization combined with mRNA expression profiling and previously characterized data on a large set of kinase inhibitors to identify kinases that are important for epithelial and mesenchymal cell migration. By profiling a selected optimal set of 32 kinase inhibitors in a panel against six cell lines, we identified cell type-specific kinases that regulate cell migration. Our discovery of several informative kinases with a previously uncharacterized role in cell migration (such as Mst and Taok family of MAPK kinases in mesenchymal cells) may represent novel targets that warrant further investigation. Target deconvolution using our ensemble approach has the potential to aid in the rational design of more potent but less toxic drug combinations.
Keywords: cancer cell migration; perturbation biology; predictive modeling; regularized regression; systems pharmacology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Step by step method: exploiting polypharmacology for drug target deconvolution.
Fig. 2.
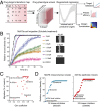
An ensemble approach of exploiting polypharmacology for kinase drug target deconvolution. (A) A schematic showing our approach of using a combination of drug-target interaction map, drug phenotypic screen, and regularized regression to exploit the polypharmacology of drugs to identify drug targets, predict efficacy, and rationally design combination therapy. (B) A real-time quantitative phenotypic method to measure cell migration using the scratch wound assay. A plot of relative wound density of Mcf10a cells treated with varying doses of Erlotinib. (Right) Representative images of cells with wound area are also shown. (C) A plot of Gini score of all 32 kinase inhibitors and their effect on relative migration in FOCUS cells. (D) Plots showing the effect of 32 kinase inhibitors on cell migration in Hs578t (mesenchymal) and Mcf10a (epithelial) breast cell lines.
Fig. 3.
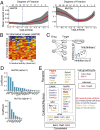
Identification of informative kinases in cell migration using elastic net regularization. (A) Plots show LOOCV error using elastic net regularization fit in HS578t and Mcf10a cell lines. The error bars represent cross-validation error plus 1 SD. The kinases identified at absolute minima (blue dashed line) were termed the most informative kinases. (B) The observed kinase activities of all informative kinases identified using elastic net regularization were affected by the panel of 32 kinase inhibitors used in our screen. A heatmap showing in vitro residual kinase activities of the 16 most informative kinases identified in Hs578t cells against 32 kinase inhibitors tested. (C) An illustration showing if two inhibitors d1 and d2 are affecting four targets K1–K4 proportionally, it would be impossible to distinguish which of the affected targets is actually responsible for the phenotype. (D) Bar graphs showing the nonzero elastic net coefficients associated with the most informative kinases (determined at α = 1) identified in Hs578t and Mcf10a cells are shown. (E) Subcellular and functional annotation of informative kinases identified in all three mesenchymal cancer cells (union of 0.7 ≤ α ≤ 1.0). Kinases with known role in cell migration are listed in bold font.
Fig. 4.
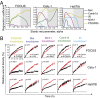
Validation of cell type-specific kinases that are determinant of mesenchymal cell migration. (A) Evolution of regression coefficients. Plots showing regression coefficients for respective kinases against value of elastic-net penalty α. Nonzero regression coefficients for kinases picked at α > 0.5 (gray region) are considered significantly informative. (B) RNAi-mediated knockdown of cell type-specific kinases in mesenchymal cancer cells. Plots showing relative wound density of indicated mesenchymal cell lines transfected with either a siRNA targeting the indicated kinase or against a scrambled control. Data are the mean of at least three independent samples and error bars indicate SEM.
Fig. 5.
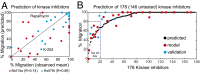
Predicting the effect of kinase inhibitors on cell migration. (A) Plot showing correlation between observed and predicted migration of HS578t and Mcf10a cells treated with 32 kinase inhibitors. Unpredictable drugs (rapamycin in Hs578t and K-252 in Mcf10a) are also indicated. (B) Informative nonzero variables determined by elastic net regularization were used to predict the efficacy of 178 (146 untested) small molecule kinase inhibitors. The black circles denote predicted values of 178 kinase inhibitors, whereas red circles denote experimental validated 32 kinase inhibitor and blue crosses denote 7 previously unseen inhibitors.
Similar articles
- Exploiting polypharmacology to dissect host kinases and kinase inhibitors that modulate endothelial barrier integrity.
Dankwa S, Dols MM, Wei L, Glennon EKK, Kain HS, Kaushansky A, Smith JD. Dankwa S, et al. Cell Chem Biol. 2021 Dec 16;28(12):1679-1692.e4. doi: 10.1016/j.chembiol.2021.06.004. Epub 2021 Jul 2. Cell Chem Biol. 2021. PMID: 34216546 Free PMC article. - Divergent Polypharmacology-Driven Cellular Activity of Structurally Similar Multi-Kinase Inhibitors through Cumulative Effects on Individual Targets.
Sumi NJ, Ctortecka C, Hu Q, Bryant AT, Fang B, Remsing Rix LL, Ayaz M, Kinose F, Welsh EA, Eschrich SA, Lawrence HR, Koomen JM, Haura EB, Rix U. Sumi NJ, et al. Cell Chem Biol. 2019 Sep 19;26(9):1240-1252.e11. doi: 10.1016/j.chembiol.2019.06.003. Epub 2019 Jun 27. Cell Chem Biol. 2019. PMID: 31257184 Free PMC article. - Rational Polypharmacology: Systematically Identifying and Engaging Multiple Drug Targets To Promote Axon Growth.
Al-Ali H, Lee DH, Danzi MC, Nassif H, Gautam P, Wennerberg K, Zuercher B, Drewry DH, Lee JK, Lemmon VP, Bixby JL. Al-Ali H, et al. ACS Chem Biol. 2015 Aug 21;10(8):1939-51. doi: 10.1021/acschembio.5b00289. Epub 2015 Jun 24. ACS Chem Biol. 2015. PMID: 26056718 Free PMC article. - Exploiting polypharmacology for improving therapeutic outcome of kinase inhibitors (KIs): An update of recent medicinal chemistry efforts.
Ma X, Lv X, Zhang J. Ma X, et al. Eur J Med Chem. 2018 Jan 1;143:449-463. doi: 10.1016/j.ejmech.2017.11.049. Epub 2017 Nov 21. Eur J Med Chem. 2018. PMID: 29202407 Review. - Polypharmacology in Drug Discovery: A Review from Systems Pharmacology Perspective.
Zhang W, Bai Y, Wang Y, Xiao W. Zhang W, et al. Curr Pharm Des. 2016;22(21):3171-81. doi: 10.2174/1381612822666160224142812. Curr Pharm Des. 2016. PMID: 26907941 Review.
Cited by
- Using machine learning to dissect host kinases required for Leishmania internalization and development.
Wei L, Barrie U, Aloisio GM, Khuong FTH, Arang N, Datta A, Kaushansky A, Wetzel DM. Wei L, et al. bioRxiv [Preprint]. 2024 Aug 4:2024.05.16.593986. doi: 10.1101/2024.05.16.593986. bioRxiv. 2024. PMID: 38798624 Free PMC article. Updated. Preprint. - Inference of drug off-target effects on cellular signaling using interactome-based deep learning.
Meimetis N, Lauffenburger DA, Nilsson A. Meimetis N, et al. iScience. 2024 Mar 14;27(4):109509. doi: 10.1016/j.isci.2024.109509. eCollection 2024 Apr 19. iScience. 2024. PMID: 38591003 Free PMC article. - Multiple receptor tyrosine kinases regulate dengue infection of hepatocytes.
Bourgeois NM, Wei L, Ho NNT, Neal ML, Seferos D, Tongogara T, Mast FD, Aitchison JD, Kaushansky A. Bourgeois NM, et al. Front Cell Infect Microbiol. 2024 Mar 22;14:1264525. doi: 10.3389/fcimb.2024.1264525. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38585651 Free PMC article. - Interrogating endothelial barrier regulation by temporally resolved kinase network generation.
Wei L, Dankwa S, Vijayan K, Smith JD, Kaushansky A. Wei L, et al. Life Sci Alliance. 2024 Mar 11;7(5):e202302522. doi: 10.26508/lsa.202302522. Print 2024 May. Life Sci Alliance. 2024. PMID: 38467420 Free PMC article. - A kinase to cytokine explorer to identify molecular regulators and potential therapeutic opportunities.
Chan M, Kang Y, Osborne S, Zager M, Gujral TS. Chan M, et al. Elife. 2024 Feb 2;12:RP91472. doi: 10.7554/eLife.91472. Elife. 2024. PMID: 38305363 Free PMC article.
References
- Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24(7):805–815. - PubMed
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. - PubMed
- Terstappen GC, Schlüpen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6(11):891–903. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources