Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi - PubMed (original) (raw)
Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi
Janelle M Hare et al. PLoS One. 2014.
Abstract
The SOS response to DNA damage that induces up to 10% of the prokaryotic genome requires RecA action to relieve LexA transcriptional repression. In Acinetobacter species, which lack LexA, the error-prone polymerase accessory UmuDAb is instead required for ddrR induction after DNA damage, suggesting it might be a LexA analog. RNA-Seq experiments defined the DNA damage transcriptome (mitomycin C-induced) of wild type, recA and umuDAb mutant strains of both A. baylyi ADP1 and A. baumannii ATCC 17978. Of the typical SOS response genes, few were differentially regulated in these species; many were repressed or absent. A striking 38.4% of all ADP1 genes, and 11.4% of all 17978 genes, were repressed under these conditions. In A. baylyi ADP1, 66 genes (2.0% of the genome), including a CRISPR/Cas system, were DNA damage-induced, and belonged to four regulons defined by differential use of recA and umuDAb. In A. baumannii ATCC 17978, however, induction of 99% of the 152 mitomycin C-induced genes depended on recA, and only 28 of these genes required umuDAb for their induction. 90% of the induced A. baumannii genes were clustered in three prophage regions, and bacteriophage particles were observed after mitomycin C treatment. These prophages encoded esvI, esvK1, and esvK2, ethanol-stimulated virulence genes previously identified in a Caenorhabditis elegans model, as well as error-prone polymerase alleles. The induction of all 17978 error-prone polymerase alleles, whether prophage-encoded or not, was recA dependent, but only these DNA polymerase V-related genes were de-repressed in the umuDAb mutant in the absence of DNA damage. These results suggest that both species possess a robust and complex DNA damage response involving both recA-dependent and recA-independent regulons, and further demonstrates that although umuDAb has a specialized role in repressing error-prone polymerases, additional regulators likely participate in these species' transcriptional response to DNA damage.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Comparison of the ADP1 and 17978 chromosomal regions indicated as putative prophages.
This to-scale diagram indicates the size and predicted gene functions from PHAST analysis within the chromosomal regions designated as putative prophages. Locations in the genome are represented by the ACIAD and A1S gene designation boundaries for ADP1 and 17978, respectively. (A) All genes represented in the ADP1 prophage region were not induced after DNA damage, and approximately one-third of the genes in this region were either hypothetical conserved phage genes, or genes of predicted phage function (with only 2 each of DNA metabolism/replication, capsid/packaging and tail genes). (B) Three chromosomal regions of induced genes in A. baumannii ATCC 17978 overlapped with three regions of the genome designated as putative prophages by our PHAST analysis and as cryptic prophages CP5, CP9 and CP14 . Genes that were not induced in this study are marked with asterisks. Previously described virulence-associated esv genes are named below each gene, and error-prone polymerase alleles are shown in bright cyan color (see legend). The specific gene function of each gene in order of its placement in each prophage is described in Table 4.
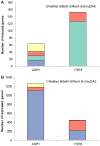
Figure 2. Distribution of regulation mechanisms for mitomycin C-induced and repressed transcriptome in ADP1 and 17978.
The absolute number of genes induced (A) or repressed (panel B) by MMC in the transcriptome of ADP1 and 17978 is shown. The designation of regulon is represented by the following terms: Neither (genes requiring neither umuDAb nor recA for regulation), Both (genes requiring both umuDAb and recA for regulation), RecA (genes requiring only recA for regulation), or UmuDAb (genes requiring only umuDAb for regulation). (A) Many more repressed genes were observed in ADP1 than 17978, with UmuDAb sufficing for this repression in most genes; 17978 repressed genes required either UmuDAb or both UmuDAb and RecA. (B) A greater number of induced genes was observed in 17978 than ADP1, and these genes required either RecA or both RecA and UmuDAb. In comparison, ADP1 induced genes belong to four regulons (Neither, Both, RecA, or UmuDAb).
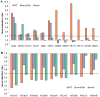
Figure 3. Mitomycin-C induced and repressed SOS genes in ADP1 and 17978 differentially require RecA and UmuDAb.
Gene induction ratios obtained from RNASeq transcriptome experiments are shown, with the induction (panel A) and repression (panel B) of canonical SOS genes. Gene names prefaced by “AD” indicate ADP1 genes; “AB” indicates 17978 genes, with A1S numbers of 17978 genes listed for umuDC alleles. The placement of the horizontal axis in each panel represents the cutoff level for a gene to be considered induced (A) or repressed (B). Bars above the horizontal axis indicate induced genes (panel A), and bars below the horizontal axis indicate repressed genes (panel B), with bars rising either below (panel A) or above (panel B) this axis to have lost their induction or repression, respectively, in the umuDAb and/or recA mutant strains. (A) Induced genes did not require umuDAb in either ADP1 or 17978, except for the category of umuDC alleles, and recA was required for all induced 17978 genes but only some induced ADP1 genes (ruvA and uvrA). (B) Repressed genes required only umuDAb in ADP1 but required both umuDAb and recA in 17978.
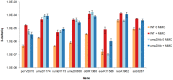
Figure 4. RT-qPCR experiments indicate that umuDAb is required for repression of error-prone polymerase components, not all DNA damage-induced genes.
Delta Cq values from RT-qPCR experiments measuring expression of selected A. baumannii ATCC 17978 genes demonstrates the repressing activity of UmuDAb only for error prone polymerase components. The expression of each gene in both wild type and umuDAb null mutant is shown, with gene identity and A1S number listed on the x axis. Each gene was assayed in one RT-qPCR experiment (plate), with error bars indicating standard error of the mean from technical triplicates of biological triplicates.
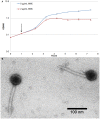
Figure 5. Mitomycin C treatment induces production of bacteriophage particles in 17978.
(A) Overnight LB cultures of 17978 cells were diluted into fresh LB medium and grown for 0.75 hours before addition of 2 μg/mL MMC. After approximately two hours of MMC treatment, the optical density leveled off and decreased slightly but continued to increase in the absence of MMC treatment. Error bars represent standard error of the mean from three independent experiments. (B) Electron micrograph of bacteriophage particles at 100,000× magnification, showing polyhedral capsid, long, flexible tail and tail fibers. Results shown are representative of three independent experiments producing and imaging bacteriophage particles.
Similar articles
- A corepressor participates in LexA-independent regulation of error-prone polymerases in Acinetobacter.
Peterson MA, Grice AN, Hare JM. Peterson MA, et al. Microbiology (Reading). 2020 Feb;166(2):212-226. doi: 10.1099/mic.0.000866. Microbiology (Reading). 2020. PMID: 31687925 Free PMC article. - UmuDAb: An Error-Prone Polymerase Accessory Homolog Whose N-Terminal Domain Is Required for Repression of DNA Damage Inducible Gene Expression in Acinetobacter baylyi.
Witkowski TA, Grice AN, Stinnett DB, Wells WK, Peterson MA, Hare JM. Witkowski TA, et al. PLoS One. 2016 Mar 24;11(3):e0152013. doi: 10.1371/journal.pone.0152013. eCollection 2016. PLoS One. 2016. PMID: 27010837 Free PMC article. - The DdrR Coregulator of the Acinetobacter baumannii Mutagenic DNA Damage Response Potentiates UmuDAb Repression of Error-Prone Polymerases.
Cook D, Flannigan MD, Candra BV, Compton KD, Hare JM. Cook D, et al. J Bacteriol. 2022 Nov 15;204(11):e0016522. doi: 10.1128/jb.00165-22. Epub 2022 Oct 4. J Bacteriol. 2022. PMID: 36194009 Free PMC article. - Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.
Jaszczur M, Bertram JG, Robinson A, van Oijen AM, Woodgate R, Cox MM, Goodman MF. Jaszczur M, et al. Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review. - Interaction of RecA mediated SOS response with bacterial persistence, biofilm formation, and host response.
Kaushik V, Tiwari M, Tiwari V. Kaushik V, et al. Int J Biol Macromol. 2022 Sep 30;217:931-943. doi: 10.1016/j.ijbiomac.2022.07.176. Epub 2022 Jul 26. Int J Biol Macromol. 2022. PMID: 35905765 Review.
Cited by
- Identification and Functional Analysis of Temperate Siphoviridae Bacteriophages of Acinetobacter baumannii.
Badawy S, Pajunen MI, Haiko J, Baka ZAM, Abou-Dobara MI, El-Sayed AKA, Skurnik M. Badawy S, et al. Viruses. 2020 May 31;12(6):604. doi: 10.3390/v12060604. Viruses. 2020. PMID: 32486497 Free PMC article. - Multiple mechanisms drive phage infection efficiency in nearly identical hosts.
Howard-Varona C, Hargreaves KR, Solonenko NE, Markillie LM, White RA 3rd, Brewer HM, Ansong C, Orr G, Adkins JN, Sullivan MB. Howard-Varona C, et al. ISME J. 2018 Jun;12(6):1605-1618. doi: 10.1038/s41396-018-0099-8. Epub 2018 Mar 22. ISME J. 2018. PMID: 29568113 Free PMC article. - The Small DdrR Protein Directly Interacts with the UmuDAb Regulator of the Mutagenic DNA Damage Response in Acinetobacter baumannii.
Pavlin A, Bajc G, Fornelos N, Browning DF, Butala M. Pavlin A, et al. J Bacteriol. 2022 Mar 15;204(3):e0060121. doi: 10.1128/jb.00601-21. Epub 2022 Feb 22. J Bacteriol. 2022. PMID: 35191762 Free PMC article. - Interspecies ecological competition rejuvenates decayed Geobacter electroactive biofilm.
Ye Y, Zhang L, Hong X, Chen M, Liu X, Zhou S. Ye Y, et al. ISME J. 2024 Jan 8;18(1):wrae118. doi: 10.1093/ismejo/wrae118. ISME J. 2024. PMID: 38916438 Free PMC article. - Alterations in gene expression of recA and umuDC in antibiotic-resistant Acinetobacter baumannii.
Ameer NAA, Dhahi MAR. Ameer NAA, et al. J Med Life. 2023 Apr;16(4):531-539. doi: 10.25122/jml-2022-0358. J Med Life. 2023. PMID: 37305826 Free PMC article.
References
- Khil PP, Camerini-Otero RD (2002) Over 1000 genes are involved in the DNA damage response of Escherichia coli . Mol Microbiol 44: 89–105. - PubMed
- Walker GC (1996) The SOS Response of Escherichia coli. Washington, D.C.: ASM Press.
- Friedberg EC (1995) DNA repair and mutagenesis. Washington, D.C.: ASM Press.
- Little JW, Mount DW (1982) The SOS regulatory system of Escherichia coli . Cell 29: 11–22. - PubMed
- Huisman O, D'Ari R (1981) An inducible DNA replication-cell division coupling mechanism in E. coli . Nature 290: 797–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 RR016481/RR/NCRR NIH HHS/United States
- P20GM103436/GM/NIGMS NIH HHS/United States
- P20 GM103436/GM/NIGMS NIH HHS/United States
- R15 GM085722/GM/NIGMS NIH HHS/United States
- R15GM085722-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources