Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein - PubMed (original) (raw)
Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein
Guifang Chen et al. Cell Host Microbe. 2014.
Abstract
Influenza A virus RNA synthesis is catalyzed by the viral polymerase comprised of the PA, PB1, and PB2 proteins. We show that the host DDX21 RNA helicase restricts influenza A virus by binding PB1 and inhibiting polymerase assembly, resulting in reduced viral RNA and protein synthesis. Later during infection, the viral NS1 protein overcomes this restriction by binding to DDX21 and displacing PB1. DDX21 binds to a region of the NS1 N-terminal domain that also participates in other critical functions. A virus mutant whose NS1 protein is unable to bind DDX21 exhibits reduced viral protein synthesis at both late and early times of infection, a phenotype converted to wild-type upon DDX21 knockdown. As sequential interaction of PB1 and NS1 with DDX21 leads to temporal regulation of viral gene expression, influenza A virus likely uses the DDX21-NS1 interaction not only to overcome restriction, but also to regulate the viral life cycle.
Copyright © 2014 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors do not have conflicts of interest.
Figures
Figure 1
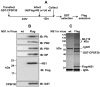
Purification of infected cell CPSF30-NS1 complexes from influenza A virus-infected cells. (A) Purification procedure. (B) Immunoblots (IBs) for the viral PA, PB1, PB2, NP and NS1 proteins and for CPSF30 in the purified CPSF30-NS1 complexes (Flag-NS1 sample) and in the control (wt NS1) sample. (C) Gel electrophoresis, followed by Coomassie blue staining, of the proteins in the purified CPSF30-NS1 complexes and the control sample. See also Table S1.
Figure 2
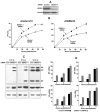
The DDX21 helicase inhibits virus replication by suppressing viral RNA synthesis at early times of infection. (A) Efficient knockdown of endogenous DDX21 in HeLa cells using DDX21-1 siRNA. Cell extracts were immunoblotted with an antibody (Ab) against DDX21 (Abcam). (B) DDX21 inhibits replication of Ud and WSN virus in HeLa cells. HeLa cells were transfected with DDX21-1 siRNA or control siRNA for 36 hours, followed by infection with 0.001 pfu/cell of Ud or WSN virus. Virus production at the indicated times after infection was determined by plaque assays in MDCK cells. The bars show the standard deviation of triplicate measurements of virus production. (C) Time course of viral protein synthesis in HeLa cells treated with DDX21-1 siRNA or control siRNA. After 36 hours of siRNA treatment, cells were infected with 2 pfu/cell of Ud virus. Cells collected at the indicated times after infection were lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) plus complete protease inhibitor (Roche). An aliquot was analyzed by IBs probed with Abs against the NS1 protein, β-actin (Cell Signaling), or a goat Ab against the major structural proteins of the Ud virus (HA, NP, and M1) provided by Robert A. Lamb (designated as Ud Ab) (Chen et al., 2007). (D) Time course of synthesis of M1 and NA mRNAs and vRNAs in HeLa cells treated with DDX21 siRNA or control siRNA. Cells were infected with 2 pfu/cell of Ud virus, and total RNA was isolated at the indicated times after infection using the TRIzol reagent. The amounts of the mRNAs and vRNAs were determined by quantitative RTPCR using the primers shown in SI. The reactions were carried out in triplicate and were normalized to the levels of β-actin mRNA amplified with β-actin specific primers. The numbers on the Y-axis correspond to the levels of the mRNA or vRNA relative to the level of the mRNA or vRNA at 1 hour postinfection in cells treated with the control siRNA. The bars show the standard deviation of the triplicate determinations. See also Figures S1 and S2.
Figure 3
Identification of the viral proteins that bind DDX21. (A) DDX21 binds NS1 and PB1 in infected cells. 293T cells were transfected for 24 hours with a plasmid expressing GST-DDX21 or GST. The cells were infected with 2 pfu/cell of Ud virus for 12 hours, and cell extracts with or without treatment with 10μg/ml of RNase A for 4 hours were GST-selected. Aliquots of the eluates were immunoblotted using the indicated Abs. Rabbit Ab against PB1 was provided by Krister Melen and Ikka Julkunen. Monoclonal Abs against PA and PB2 were provided by Juan Ortin (Ochoa et al., 1995). (B) DDX21 binds the PB1 protein of several influenza A virus strains in uninfected cells. 293T cells were transfected for 48 hours with a plasmid expressing the PB1 protein from the indicated influenza A virus strain and a plasmid expressing GST-DDX21 or GST. Cell extracts were prepared by treating cells with a buffer containing 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.5% Nonidet P-40, plus complete protease inhibitor, followed by rotation of the extracts for 30 minutes at 4°C. After clarification by low speed centrifugation at 4°C. the extracts with and without RNase A treatment (10μg/ml) were GST selected using glutathione magnet beads (Pierce), and the eluates were immunoblotted with PB1 Ab. Also see Figures S3 and S4.
Figure 4
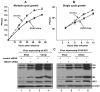
DDX21 inhibits viral RNA synthesis by binding PB1 and inhibiting formation of the viral polymerase. (A) Effect of DDX21 on viral RNA synthesis measured in dual luciferase reporter assays carried out as described in the text. The Firefly/Renilla luciferase ratio, which measures luciferase mRNA synthesis, was determined in the absence and presence of the indicated amounts of the transfected DDX21 plasmid. The ratios in the presence of the DDX21 plasmid were normalized to the ratio in the absence of DDX21. The assays were carried out in triplicate. The bars show the standard deviation of the triplicate determinations. Quantitative RT-PCR was carried out in triplicate to verify that this ratio accurately reflects the amount of luciferase mRNA produced (right lower panel). The Firefly mRNA levels in cells transfected with the DDX21 plasmid was normalized to the Firefly mRNA level in cells transfected with the empty plasmid (0μg DDX21). The bars show the range of the triplicate results. The levels of the viral proteins in the absence and presence (1μg) of the DDX21 plasmid were measured by IBs to establish that DDX21 did not affect the synthesis of the viral proteins (left lower panel). (B) DDX21 lacking helicase activity maintains the ability to inhibit viral RNA synthesis. The DDX21 plasmid (1μg) that was co-transfected in the luciferase reporter assay expressed either wt DDX21 or DDX21 in which the DEAD sequence was changed to AAAD. (C) DDX21 inhibits the formation of the viral polymerase. 293T cells were co-tranfected for 48 hours with plasmids expressing TAP-tagged PB1, PA, and PB2, and with a plasmid expressing DDX21 or an empty vector (EV). Cell extracts prepared as described in the legend of Figure 2B were TAP-selected, and the eluates were immunoblotted with the indicated Abs. ImageJ software (NIH) was used to quantitate the PB1, PA and PB2 IBs. p values calculated via a one-tailed Student’s t-test are shown. See also Figure S5.
Figure 5
NS1 protein binds DDX21 and displaces PB1 from DDX21. (A) DDX21 binds the NS1 protein in uninfected cells. 293T cells were transfected for 48 hours with a plasmid expressing the Ud NS1 protein and a plasmid expressing GST-DDX21 or GST. Cell extracts prepared as described in the legend of Figure 3B, with and without RNase A treatment, were GST selected, and the eluates were immunoblotted with NS1 Ab. (B) NS1 displaces PB1 from binding DDX21. 293T cells were transfected for 48 hours with the indicated amounts of plasmids expressing GST-DDX21, 3xFlag-WSN NS1, and Ud PB1. Cell extracts treated with RNase A were GST selected, and the amounts of PB1 and NS1 in the eluates were measured by IBs probed with PB1 or Flag Ab, respectively.
Figure 6
Identification of the amino acids in the NS1 RBD that are required for binding DDX21. (A) Bacteria-expressed GST and the indicated GST-tagged NS1 proteins were bound to glutathione magnetic beads, which were then incubated in the presence of RNase A for 4 hours with 293T cell extracts containing plasmid-expressed Flag-DDX21. The proteins eluted from the beads were analyzed by an IB probed with Flag Ab (top panel) and by Coomassie blue staining (bottom panel). (B) The structure of the RBD showing the amino acids (R37/R37’, R38/R38’, K41/K41’, R44/R44’, K70/K70’) that were replaced with alanines. The RBD structure was generated from PBD id 1NS1. (C) Bacteria extracts containing GST, GST-NS1 (1-73), or GST-NS1 (1-73) containing the indicated amino acid substitutions were bound to glutathione magnetic beads, which were then Incubated in the presence of RNase A with 293T containing plasmid-expressed Flag-DDX21. The eluted proteins were analyzed by an IB probed with Flag Ab and by Coomassie blue staining. (D) 293T cells were co-transfected for 48 hours with plasmids expressing GST-DDX21, PB1, and either wt or 41/44 NS1 protein. Cell extracts were RNase A treated and GST selected, and the eluates were immunoblotted with PB1 or Flag Ab. (E) The dual luciferase assay for viral RNA synthesis was carried in triplicate out as described in the text. Where indicated, 1μg of a plasmid expressing DDX21, and 1μg of a plasmid expressing either wt or 41/44 NS1 protein were added. The Firefly/Renilla ratio was normalized to the ratio in the absence of DDX21. The levels of the wt and 41/44 NS1 proteins, DDX21 and β-actin were determined by IBs.
Figure 7
The NS1 protein counteracts endogenous DDX21 antiviral activity during virus infection. (A) Calu3 cells were infected with 0.001 pfu/cell of wt or 41/44 virus. Virus production at the indicated times after infection was determined by plaque assays. The bars show the standard deviation of triplicate measurements of virus production. (B) Calu3 cells were infected with 5 pfu/cell of wt or 41/44 virus. The bars show the standard deviation of triplicate measurements of virus production. (C) HeLa cells were transfected with a DDX21 siRNA or a control siRNA for 36 hours, followed by infection with 2 pfu/cell of wt or 41/44 virus. At 9 and 12 hours after infection, cell extracts were immunoblotted with Ud, NS1, or β-actin Ab. See also Figure S6.
Similar articles
- Quantitative proteomic analysis of the influenza A virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor.
Tawaratsumida K, Phan V, Hrincius ER, High AA, Webby R, Redecke V, Häcker H. Tawaratsumida K, et al. J Virol. 2014 Aug;88(16):9038-48. doi: 10.1128/JVI.00830-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899174 Free PMC article. - Phosphorylation of Influenza A Virus NS1 at Serine 205 Mediates Its Viral Polymerase-Enhancing Function.
Patil A, Anhlan D, Ferrando V, Mecate-Zambrano A, Mellmann A, Wixler V, Boergeling Y, Ludwig S. Patil A, et al. J Virol. 2021 Feb 24;95(6):e02369-20. doi: 10.1128/JVI.02369-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33408177 Free PMC article. - The Short Form of the Zinc Finger Antiviral Protein Inhibits Influenza A Virus Protein Expression and Is Antagonized by the Virus-Encoded NS1.
Tang Q, Wang X, Gao G. Tang Q, et al. J Virol. 2017 Jan 3;91(2):e01909-16. doi: 10.1128/JVI.01909-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27807230 Free PMC article. - Host Shutoff in Influenza A Virus: Many Means to an End.
Levene RE, Gaglia MM. Levene RE, et al. Viruses. 2018 Sep 5;10(9):475. doi: 10.3390/v10090475. Viruses. 2018. PMID: 30189604 Free PMC article. Review. - The NS1 protein: a multitasking virulence factor.
Ayllon J, García-Sastre A. Ayllon J, et al. Curr Top Microbiol Immunol. 2015;386:73-107. doi: 10.1007/82_2014_400. Curr Top Microbiol Immunol. 2015. PMID: 25007846 Review.
Cited by
- Influenza Virus Host Restriction Factors: The ISGs and Non-ISGs.
Husain M. Husain M. Pathogens. 2024 Jan 29;13(2):127. doi: 10.3390/pathogens13020127. Pathogens. 2024. PMID: 38392865 Free PMC article. Review. - Multiomics approach reveals the ubiquitination-specific processes hijacked by SARS-CoV-2.
Xu G, Wu Y, Xiao T, Qi F, Fan L, Zhang S, Zhou J, He Y, Gao X, Zeng H, Li Y, Zhang Z. Xu G, et al. Signal Transduct Target Ther. 2022 Sep 7;7(1):312. doi: 10.1038/s41392-022-01156-y. Signal Transduct Target Ther. 2022. PMID: 36071039 Free PMC article. - RuvB-Like Protein 2 Interacts with the NS1 Protein of Influenza A Virus and Affects Apoptosis That Is Counterbalanced by Type I Interferons.
Wang Y, Zhou J, Mackintosh SG, Du Y. Wang Y, et al. Viruses. 2021 May 31;13(6):1038. doi: 10.3390/v13061038. Viruses. 2021. PMID: 34072766 Free PMC article. - The mitochondrial carrier homolog 2 is involved in down-regulation of influenza A virus replication.
Ulupinar P, Çağlayan E, Rayaman E, Nagata K, Turan K. Ulupinar P, et al. Mol Biol Rep. 2024 May 10;51(1):642. doi: 10.1007/s11033-024-09584-5. Mol Biol Rep. 2024. PMID: 38727866 - Unveiling the role of host kinases at different steps of influenza A virus life cycle.
Dey S, Mondal A. Dey S, et al. J Virol. 2024 Jan 23;98(1):e0119223. doi: 10.1128/jvi.01192-23. Epub 2024 Jan 4. J Virol. 2024. PMID: 38174932 Free PMC article. Review.
References
- Bradel-Tretheway BG, Mattiacio JL, Krasnoselsky A, Stevenson C, Purdy D, Dewhurst S, Katze MG. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J Virol. 2011;85:8569–8581. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous