BEAST 2: a software platform for Bayesian evolutionary analysis - PubMed (original) (raw)
BEAST 2: a software platform for Bayesian evolutionary analysis
Remco Bouckaert et al. PLoS Comput Biol. 2014.
Abstract
We present a new open source, extensible and flexible software platform for Bayesian evolutionary analysis called BEAST 2. This software platform is a re-design of the popular BEAST 1 platform to correct structural deficiencies that became evident as the BEAST 1 software evolved. Key among those deficiencies was the lack of post-deployment extensibility. BEAST 2 now has a fully developed package management system that allows third party developers to write additional functionality that can be directly installed to the BEAST 2 analysis platform via a package manager without requiring a new software release of the platform. This package architecture is showcased with a number of recently published new models encompassing birth-death-sampling tree priors, phylodynamics and model averaging for substitution models and site partitioning. A second major improvement is the ability to read/write the entire state of the MCMC chain to/from disk allowing it to be easily shared between multiple instances of the BEAST software. This facilitates checkpointing and better support for multi-processor and high-end computing extensions. Finally, the functionality in new packages can be easily added to the user interface (BEAUti 2) by a simple XML template-based mechanism because BEAST 2 has been re-designed to provide greater integration between the analysis engine and the user interface so that, for example BEAST and BEAUti use exactly the same XML file format.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
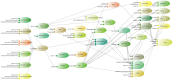
Figure 1. A complete model with six sequences, HKY substitution model, strict clock, Yule tree prior, a number of operators, mainly on the tree, and a few loggers to produce output to a trace file, a tree log and screen output.
Note the explicit existence of a State object.
Figure 2. Results from an SDPM2 partition and substitution model averaging analysis of mtDNA from 7 species of primate (data from [22]).
(a) The posterior probability distribution of the substitution model given a site. The grid cell at row i and column j represents the posterior probability of model i at site j. The darker the shade, the higher the posterior probability. It appears that this data favours models that accommodate a difference in transition and transversion rates and in nucleotide base frequencies. Of those three models, the simplest version (HKY85) is generally preferred. (b) There are four biological partitions in this data set: the three codon positions and a tRNA region. Conditioned on three rate categories (which has the highest posterior probability), the mean posterior proportion of sites in each category for each biological partition is plotted. The categories with faster rates are closer to the top of the bar.
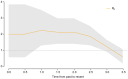
Figure 3. Bayesian estimate of the effective reproduction number Re over time (from simulated data).
Towards the end of the 3.5 years of the underlying epidemic process, Re decreases below 1, which indicates a declining epidemic.
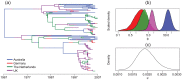
Figure 4. Results from a structured coalescent analysis of a spatially-annotated HCV data set using the MultiTypeTree package.
These include (a) a typical tree drawn from the posterior (edge colours represent lineage locations), (b) posterior distributions of θ = Neg for each of the spatially-localized sub-populations and (c) the posterior distribution for the molecular clock rate μ 0.
Similar articles
- Bayesian phylogenetics with BEAUti and the BEAST 1.7.
Drummond AJ, Suchard MA, Xie D, Rambaut A. Drummond AJ, et al. Mol Biol Evol. 2012 Aug;29(8):1969-73. doi: 10.1093/molbev/mss075. Epub 2012 Feb 25. Mol Biol Evol. 2012. PMID: 22367748 Free PMC article. - BEAST: Bayesian evolutionary analysis by sampling trees.
Drummond AJ, Rambaut A. Drummond AJ, et al. BMC Evol Biol. 2007 Nov 8;7:214. doi: 10.1186/1471-2148-7-214. BMC Evol Biol. 2007. PMID: 17996036 Free PMC article. - BEASTling: A software tool for linguistic phylogenetics using BEAST 2.
Maurits L, Forkel R, Kaiping GA, Atkinson QD. Maurits L, et al. PLoS One. 2017 Aug 10;12(8):e0180908. doi: 10.1371/journal.pone.0180908. eCollection 2017. PLoS One. 2017. PMID: 28796784 Free PMC article. - πBUSS: a parallel BEAST/BEAGLE utility for sequence simulation under complex evolutionary scenarios.
Bielejec F, Lemey P, Carvalho LM, Baele G, Rambaut A, Suchard MA. Bielejec F, et al. BMC Bioinformatics. 2014 May 7;15:133. doi: 10.1186/1471-2105-15-133. BMC Bioinformatics. 2014. PMID: 24885610 Free PMC article. - The State of Software for Evolutionary Biology.
Darriba D, Flouri T, Stamatakis A. Darriba D, et al. Mol Biol Evol. 2018 May 1;35(5):1037-1046. doi: 10.1093/molbev/msy014. Mol Biol Evol. 2018. PMID: 29385525 Free PMC article. Review.
Cited by
- Staphylococcus aureus ST59: Concurrent but Separate Evolution of North American and East Asian Lineages.
McClure JA, Lakhundi S, Niazy A, Dong G, Obasuyi O, Gordon P, Chen S, Conly JM, Zhang K. McClure JA, et al. Front Microbiol. 2021 Feb 10;12:631845. doi: 10.3389/fmicb.2021.631845. eCollection 2021. Front Microbiol. 2021. PMID: 33643261 Free PMC article. - Whole genome sequencing of Streptomyces actuosus ISP-5337, Streptomyces sioyaensis B-5408, and Actinospica acidiphila B-2296 reveals secondary metabolomes with antibiotic potential.
Majer HM, Ehrlich RL, Ahmed A, Earl JP, Ehrlich GD, Beld J. Majer HM, et al. Biotechnol Rep (Amst). 2021 Feb 9;29:e00596. doi: 10.1016/j.btre.2021.e00596. eCollection 2021 Mar. Biotechnol Rep (Amst). 2021. PMID: 33643857 Free PMC article. - Natural infection of parvovirus in wild fishing cats (Prionailurus viverrinus) reveals extant viral localization in kidneys.
Piewbang C, Wardhani SW, Chanseanroj J, Yostawonkul J, Boonrungsiman S, Saengkrit N, Kongmakee P, Banlunara W, Poovorawan Y, Kasantikul T, Techangamsuwan S. Piewbang C, et al. PLoS One. 2021 Mar 2;16(3):e0247266. doi: 10.1371/journal.pone.0247266. eCollection 2021. PLoS One. 2021. PMID: 33651823 Free PMC article. - Insights into the molecular diversity of Plasmodium vivax merozoite surface protein-3γ (pvmsp3γ), a polymorphic member in the msp3 multi-gene family.
Kuamsab N, Putaporntip C, Pattanawong U, Jongwutiwes S. Kuamsab N, et al. Sci Rep. 2020 Jul 3;10(1):10977. doi: 10.1038/s41598-020-67222-z. Sci Rep. 2020. PMID: 32620822 Free PMC article. - Shallow Whole-Genome Sequencing of Aedes japonicus and Aedes koreicus from Italy and an Updated Picture of Their Evolution Based on Mitogenomics and Barcoding.
Zadra N, Tatti A, Silverj A, Piccinno R, Devilliers J, Lewis C, Arnoldi D, Montarsi F, Escuer P, Fusco G, De Sanctis V, Feuda R, Sánchez-Gracia A, Rizzoli A, Rota-Stabelli O. Zadra N, et al. Insects. 2023 Nov 23;14(12):904. doi: 10.3390/insects14120904. Insects. 2023. PMID: 38132578 Free PMC article.
References
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–92. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources