ProteomeXchange provides globally coordinated proteomics data submission and dissemination - PubMed (original) (raw)
Eric W Deutsch 2, Rui Wang 3, Attila Csordas 3, Florian Reisinger 3, Daniel Ríos 3, José A Dianes 3, Zhi Sun 4, Terry Farrah 4, Nuno Bandeira 5, Pierre-Alain Binz 6, Ioannis Xenarios 7, Martin Eisenacher 8, Gerhard Mayer 8, Laurent Gatto 9, Alex Campos 10, Robert J Chalkley 11, Hans-Joachim Kraus 12, Juan Pablo Albar 13, Salvador Martinez-Bartolomé 13, Rolf Apweiler 3, Gilbert S Omenn 14, Lennart Martens 15, Andrew R Jones 16, Henning Hermjakob 3
Affiliations
- PMID: 24727771
- PMCID: PMC3986813
- DOI: 10.1038/nbt.2839
ProteomeXchange provides globally coordinated proteomics data submission and dissemination
Juan A Vizcaíno et al. Nat Biotechnol. 2014 Mar.
No abstract available
Conflict of interest statement
Competing financial interests
The authors have no competing financial or commercial interests.
Figures
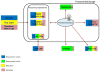
Figure 1
Representation of the ProteomeXchange workflow for MS/MS and SRM data. *Raw data represents mass spectrometer output files.
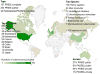
Figure 2
Summary of the main metrics of ProteomeXchange submissions (by August 2013). The number of data sets is indicated for submission type, data access status and for the top species and countries represented.
Similar articles
- Using the PRIDE Database and ProteomeXchange for Submitting and Accessing Public Proteomics Datasets.
Jarnuczak AF, Vizcaíno JA. Jarnuczak AF, et al. Curr Protoc Bioinformatics. 2017 Sep 13;59:13.31.1-13.31.12. doi: 10.1002/cpbi.30. Curr Protoc Bioinformatics. 2017. PMID: 28902400 - PRIDE: Data submission and analysis.
Vizcaíno JA, Reisinger F, Côté R, Martens L. Vizcaíno JA, et al. Curr Protoc Protein Sci. 2010 Apr;Chapter 25:25.4.1-25.4.11. doi: 10.1002/0471140864.ps2504s60. Curr Protoc Protein Sci. 2010. PMID: 20393974 - The jPOST Repository as a Public Data Repository for Shotgun Proteomics.
Watanabe Y, Yoshizawa AC, Ishihama Y, Okuda S. Watanabe Y, et al. Methods Mol Biol. 2021;2259:309-322. doi: 10.1007/978-1-0716-1178-4_20. Methods Mol Biol. 2021. PMID: 33687724 - Making proteomics data accessible and reusable: current state of proteomics databases and repositories.
Perez-Riverol Y, Alpi E, Wang R, Hermjakob H, Vizcaíno JA. Perez-Riverol Y, et al. Proteomics. 2015 Mar;15(5-6):930-49. doi: 10.1002/pmic.201400302. Proteomics. 2015. PMID: 25158685 Free PMC article. Review. - Proteomic repository data submission, dissemination, and reuse: key messages.
Perez-Riverol Y. Perez-Riverol Y. Expert Rev Proteomics. 2022 Jul-Dec;19(7-12):297-310. doi: 10.1080/14789450.2022.2160324. Epub 2022 Dec 26. Expert Rev Proteomics. 2022. PMID: 36529941 Free PMC article. Review.
Cited by
- Dynamic rewiring of the human interactome by interferon signaling.
Kerr CH, Skinnider MA, Andrews DDT, Madero AM, Chan QWT, Stacey RG, Stoynov N, Jan E, Foster LJ. Kerr CH, et al. Genome Biol. 2020 Jun 15;21(1):140. doi: 10.1186/s13059-020-02050-y. Genome Biol. 2020. PMID: 32539747 Free PMC article. - An improved protocol to study the plant cell wall proteome.
Printz B, Dos Santos Morais R, Wienkoop S, Sergeant K, Lutts S, Hausman JF, Renaut J. Printz B, et al. Front Plant Sci. 2015 Apr 10;6:237. doi: 10.3389/fpls.2015.00237. eCollection 2015. Front Plant Sci. 2015. PMID: 25914713 Free PMC article. - Combination of Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and Substrate Trapping for the Detection of Transient Protein Interactions.
Duda JM, Thomas SN. Duda JM, et al. Methods Mol Biol. 2023;2603:219-234. doi: 10.1007/978-1-0716-2863-8_18. Methods Mol Biol. 2023. PMID: 36370283 Free PMC article. - A proteomic analysis of chondrogenic, osteogenic and tenogenic constructs from ageing mesenchymal stem cells.
Peffers MJ, Collins J, Loughlin J, Proctor C, Clegg PD. Peffers MJ, et al. Stem Cell Res Ther. 2016 Sep 14;7(1):133. doi: 10.1186/s13287-016-0384-2. Stem Cell Res Ther. 2016. PMID: 27624072 Free PMC article. - Mining Missing Membrane Proteins by High-pH Reverse-Phase StageTip Fractionation and Multiple Reaction Monitoring Mass Spectrometry.
Kitata RB, Dimayacyac-Esleta BR, Choong WK, Tsai CF, Lin TD, Tsou CC, Weng SH, Chen YJ, Yang PC, Arco SD, Nesvizhskii AI, Sung TY, Chen YJ. Kitata RB, et al. J Proteome Res. 2015 Sep 4;14(9):3658-69. doi: 10.1021/acs.jproteome.5b00477. Epub 2015 Aug 6. J Proteome Res. 2015. PMID: 26202522 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- P41 GM103481/GM/NIGMS NIH HHS/United States
- WT085949MA/WT_/Wellcome Trust/United Kingdom
- P41 GM103484/GM/NIGMS NIH HHS/United States
- R01 GM087221/GM/NIGMS NIH HHS/United States
- BB/I024204/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P30U54ES017885/ES/NIEHS NIH HHS/United States
- UL1RR24986/RR/NCRR NIH HHS/United States
- BB/I00095X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 085949/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources