Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data - PubMed (original) (raw)
. 2014 Sep 1;23(17):4710-20.
doi: 10.1093/hmg/ddu174. Epub 2014 Apr 11.
Collaborators, Affiliations
- PMID: 24728037
- PMCID: PMC4119411
- DOI: 10.1093/hmg/ddu174
Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data
Guo-Bo Chen et al. Hum Mol Genet. 2014.
Abstract
As custom arrays are cheaper than generic GWAS arrays, larger sample size is achievable for gene discovery. Custom arrays can tag more variants through denser genotyping of SNPs at associated loci, but at the cost of losing genome-wide coverage. Balancing this trade-off is important for maximizing experimental designs. We quantified both the gain in captured SNP-heritability at known candidate regions and the loss due to imperfect genome-wide coverage for inflammatory bowel disease using immunochip (iChip) and imputed GWAS data on 61,251 and 38.550 samples, respectively. For Crohn's disease (CD), the iChip and GWAS data explained 19 and 26% of variation in liability, respectively, and SNPs in the densely genotyped iChip regions explained 13% of the SNP-heritability for both the iChip and GWAS data. For ulcerative colitis (UC), the iChip and GWAS data explained 15 and 19% of variation in liability, respectively, and the dense iChip regions explained 10 and 9% of the SNP-heritability in the iChip and the GWAS data. From bivariate analyses, estimates of the genetic correlation in risk between CD and UC were 0.75 (SE 0.017) and 0.62 (SE 0.042) for the iChip and GWAS data, respectively. We also quantified the SNP-heritability of genomic regions that did or did not contain the previous 163 GWAS hits for CD and UC, and SNP-heritability of the overlapping loci between the densely genotyped iChip regions and the 163 GWAS hits. For both diseases, over different genomic partitioning, the densely genotyped regions on the iChip tagged at least as much variation in liability as in the corresponding regions in the GWAS data, however a certain amount of tagged SNP-heritability in the GWAS data was lost using the iChip due to the low coverage at unselected regions. These results imply that custom arrays with a GWAS backbone will facilitate more gene discovery, both at associated and novel loci.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
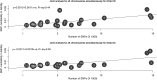
Figure 1.
Increase in the number of GWAS-significant signals for total sample size and the number of cases. Both the axes are on a logarithmic scale. The source of the data is in
Supplementary Material, Table S1
.
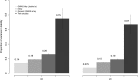
Figure 2.
The comprehensive estimation of the explained variance in liability scale for CD and UC. For each disease, from left to right are the estimates from GWAS hits (12), iChip, generic GWAS array data, and twin studies pooling four published twins cohorts together (
Supplementary Material, Note
).
Figure 3.
Partitioning of genetic variation in liability to CD and UC across chromosomes. The iChip sample was split into two subsets, and the averaged heritability was used in this figure.
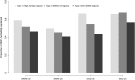
Figure 4.
Proportion of the genetic variation in liability to CD and UC explained by the selected regions across three types of partitioning of the genome.
Similar articles
- Performance of risk prediction for inflammatory bowel disease based on genotyping platform and genomic risk score method.
Chen GB, Lee SH, Montgomery GW, Wray NR, Visscher PM, Gearry RB, Lawrance IC, Andrews JM, Bampton P, Mahy G, Bell S, Walsh A, Connor S, Sparrow M, Bowdler LM, Simms LA, Krishnaprasad K; International IBD Genetics Consortium; Radford-Smith GL, Moser G. Chen GB, et al. BMC Med Genet. 2017 Aug 29;18(1):94. doi: 10.1186/s12881-017-0451-2. BMC Med Genet. 2017. PMID: 28851283 Free PMC article. - Quantifying missing heritability at known GWAS loci.
Gusev A, Bhatia G, Zaitlen N, Vilhjalmsson BJ, Diogo D, Stahl EA, Gregersen PK, Worthington J, Klareskog L, Raychaudhuri S, Plenge RM, Pasaniuc B, Price AL. Gusev A, et al. PLoS Genet. 2013;9(12):e1003993. doi: 10.1371/journal.pgen.1003993. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385918 Free PMC article. - Genetic overlap between inflammatory bowel disease and iridocyclitis: insights from a genome-wide association study in a European population.
Liao W, Luo Q, Zhang L, Wang H, Ge W, Wang J, Zuo Z. Liao W, et al. BMC Genom Data. 2024 Oct 29;25(1):92. doi: 10.1186/s12863-024-01274-2. BMC Genom Data. 2024. PMID: 39472800 Free PMC article. - Heritability in inflammatory bowel disease: from the first twin study to genome-wide association studies.
Gordon H, Trier Moller F, Andersen V, Harbord M. Gordon H, et al. Inflamm Bowel Dis. 2015 Jun;21(6):1428-34. doi: 10.1097/MIB.0000000000000393. Inflamm Bowel Dis. 2015. PMID: 25895112 Free PMC article. Review. - The genetics of non-monogenic IBD.
Jans D, Cleynen I. Jans D, et al. Hum Genet. 2023 May;142(5):669-682. doi: 10.1007/s00439-023-02521-9. Epub 2023 Jan 31. Hum Genet. 2023. PMID: 36720734 Review.
Cited by
- Polygenic scores and their applications in kidney disease.
Khan A, Kiryluk K. Khan A, et al. Nat Rev Nephrol. 2024 Sep 13. doi: 10.1038/s41581-024-00886-2. Online ahead of print. Nat Rev Nephrol. 2024. PMID: 39271761 Review. - The Effect of Protein Nutritional Support on Inflammatory Bowel Disease and Its Potential Mechanisms.
Li Q, Wang J. Li Q, et al. Nutrients. 2024 Jul 17;16(14):2302. doi: 10.3390/nu16142302. Nutrients. 2024. PMID: 39064745 Free PMC article. Review. - A Polygenic Risk Analysis for Identifying Ulcerative Colitis Patients with European Ancestry.
Liu L, Wu Y, Li Y, Li M. Liu L, et al. Genes (Basel). 2024 May 25;15(6):684. doi: 10.3390/genes15060684. Genes (Basel). 2024. PMID: 38927620 Free PMC article. - A broken network of susceptibility genes in the monocytes of Crohn's disease patients.
Liu H, Guan L, Su X, Zhao L, Shu Q, Zhang J. Liu H, et al. Life Sci Alliance. 2024 Jun 26;7(9):e202302394. doi: 10.26508/lsa.202302394. Print 2024 Sep. Life Sci Alliance. 2024. PMID: 38925865 Free PMC article. - Investigating shared genetic architecture between inflammatory bowel diseases and primary biliary cholangitis.
Huang W, Jiang R, Li S, Zeng R, Li Y, Zhang Y, Tong S, Lyu Y, Wang J, Lian Q, Leung FW, Luo R, Sha W, Chen H. Huang W, et al. JHEP Rep. 2024 Feb 12;6(6):101037. doi: 10.1016/j.jhepr.2024.101037. eCollection 2024 Jun. JHEP Rep. 2024. PMID: 38721342 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- G0800675/MRC_/Medical Research Council/United Kingdom
- DE130100614/DE/NIDCR NIH HHS/United States
- P01 GM099568/GM/NIGMS NIH HHS/United States
- MRF_C0482/MRF/MRF/United Kingdom
- ETM/137/CSO_/Chief Scientist Office/United Kingdom
- ETM/75/CSO_/Chief Scientist Office/United Kingdom
- CZB/4/540/CSO_/Chief Scientist Office/United Kingdom
- MH100141/MH/NIMH NIH HHS/United States
- GM075091/GM/NIGMS NIH HHS/United States
- G0600329/MRC_/Medical Research Council/United Kingdom
- GM099568/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials