Crystal structure of yeast DNA polymerase ε catalytic domain - PubMed (original) (raw)
Crystal structure of yeast DNA polymerase ε catalytic domain
Rinku Jain et al. PLoS One. 2014.
Abstract
DNA polymerase ε (Polε) is a multi-subunit polymerase that contributes to genomic stability via its roles in leading strand replication and the repair of damaged DNA. Here we report the ternary structure of the Polε catalytic subunit (Pol2) bound to a nascent G:C base pair (Pol2G:C). Pol2G:C has a typical B-family polymerase fold and embraces the template-primer duplex with the palm, fingers, thumb and exonuclease domains. The overall arrangement of domains is similar to the structure of Pol2T:A reported recently, but there are notable differences in their polymerase and exonuclease active sites. In particular, we observe Ca2+ ions at both positions A and B in the polymerase active site and also observe a Ca2+ at position B of the exonuclease site. We find that the contacts to the nascent G:C base pair in the Pol2G:C structure are maintained in the Pol2T:A structure and reflect the comparable fidelity of Pol2 for nascent purine-pyrimidine and pyrimidine-purine base pairs. We note that unlike that of Pol3, the shape of the nascent base pair binding pocket in Pol2 is modulated from the major grove side by the presence of Tyr431. Together with Pol2T:A, our results provide a framework for understanding the structural basis of high fidelity DNA synthesis by Pol2.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
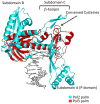
Figure 1. Structure of Pol2-DNA-dCTP ternary complex.
Pol2 palm, fingers, thumb, exonuclease and N-terminal domains are shown in cyan, yellow, orange, magenta and blue respectively. DNA is in gray, and the templating G and incoming dCTP are in red. The polymerase (Pol) and exonuclease (Exo) active sites are labeled.
Figure 2. Comparison between the palm domains of Pol2 and Pol3.
Pol2 palm domain is in cyan and the Pol3 palm domain is in red. DNA is in gray. Structural insertions in the Pol2 palm domain are highlighted. The Pol2 palm domain is more elaborate and contains insertions that form three subdomains – A, B, and C.
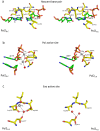
Figure 3. Comparison between the Pol2G:C and Pol2T:A polymerase and exonuclease active sites.
(a) The nascent base pair binding pocket is shaped by residues Val825 (not shown for clarity), Asn828, Ser829, Ty831 and Gly832 from the fingers domain, and by Tyr645 from the palm domain. Tyr431 approaches the incoming nucleotide from the major groove side. Contacts to the G:C and T:A base pairs are interchangeable in the two structures. (b) The polymerase active site is characterized by acidic residues Asp640 and Asp877. The Pol2G:C (this work) structure has two Ca2+ ions (gray spheres) at positions A and B in the polymerase active site. Pol2T:A structure was crystallized with one Mg2+ ion in the active site. (c) Exonuclease active site in Pol2GC (left) and Pol2TA (right). Ca2+ ion at position B of Pol2G:C is shown as gray sphere and is coordinated by Asp290, and a water molecule (red sphere). The atom at position A was modeled as water due to its close proximity to the metal ion at position B. In the Pol2T:A structure, the exonuclease catalytic residues (Asp290 and Glu292) were mutated to alanines and there are no bound metal ions.
Similar articles
- Checkpoint-mediated DNA polymerase ε exonuclease activity curbing counteracts resection-driven fork collapse.
Pellicanò G, Al Mamun M, Jurado-Santiago D, Villa-Hernández S, Yin X, Giannattasio M, Lanz MC, Smolka MB, Yeeles J, Shirahige K, García-Díaz M, Bermejo R. Pellicanò G, et al. Mol Cell. 2021 Jul 1;81(13):2778-2792.e4. doi: 10.1016/j.molcel.2021.04.006. Epub 2021 Apr 30. Mol Cell. 2021. PMID: 33932350 Free PMC article. - An iron-sulfur cluster in the polymerase domain of yeast DNA polymerase ε.
Jain R, Vanamee ES, Dzikovski BG, Buku A, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Jain R, et al. J Mol Biol. 2014 Jan 23;426(2):301-8. doi: 10.1016/j.jmb.2013.10.015. Epub 2013 Oct 19. J Mol Biol. 2014. PMID: 24144619 Free PMC article. - Structural evidence for an essential Fe-S cluster in the catalytic core domain of DNA polymerase ϵ.
Ter Beek J, Parkash V, Bylund GO, Osterman P, Sauer-Eriksson AE, Johansson E. Ter Beek J, et al. Nucleic Acids Res. 2019 Jun 20;47(11):5712-5722. doi: 10.1093/nar/gkz248. Nucleic Acids Res. 2019. PMID: 30968138 Free PMC article. - Fidelity of DNA polymerase epsilon holoenzyme from budding yeast Saccharomyces cerevisiae.
Shimizu K, Hashimoto K, Kirchner JM, Nakai W, Nishikawa H, Resnick MA, Sugino A. Shimizu K, et al. J Biol Chem. 2002 Oct 4;277(40):37422-9. doi: 10.1074/jbc.M204476200. Epub 2002 Jul 17. J Biol Chem. 2002. PMID: 12124389 - The evolving tale of Pol2 function.
Gallitto M, Zhang Z. Gallitto M, et al. Genes Dev. 2023 Feb 1;37(3-4):72-73. doi: 10.1101/gad.350527.123. Epub 2023 Feb 22. Genes Dev. 2023. PMID: 36813532 Free PMC article. Review.
Cited by
- Human DNA polymerase α in binary complex with a DNA:DNA template-primer.
Coloma J, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Coloma J, et al. Sci Rep. 2016 Apr 1;6:23784. doi: 10.1038/srep23784. Sci Rep. 2016. PMID: 27032819 Free PMC article. - POLE mutations in families predisposed to cutaneous melanoma.
Aoude LG, Heitzer E, Johansson P, Gartside M, Wadt K, Pritchard AL, Palmer JM, Symmons J, Gerdes AM, Montgomery GW, Martin NG, Tomlinson I, Kearsey S, Hayward NK. Aoude LG, et al. Fam Cancer. 2015 Dec;14(4):621-8. doi: 10.1007/s10689-015-9826-8. Fam Cancer. 2015. PMID: 26251183 - Checkpoint-mediated DNA polymerase ε exonuclease activity curbing counteracts resection-driven fork collapse.
Pellicanò G, Al Mamun M, Jurado-Santiago D, Villa-Hernández S, Yin X, Giannattasio M, Lanz MC, Smolka MB, Yeeles J, Shirahige K, García-Díaz M, Bermejo R. Pellicanò G, et al. Mol Cell. 2021 Jul 1;81(13):2778-2792.e4. doi: 10.1016/j.molcel.2021.04.006. Epub 2021 Apr 30. Mol Cell. 2021. PMID: 33932350 Free PMC article. - Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other.
Nandakumar D, Pandey M, Patel SS. Nandakumar D, et al. Elife. 2015 May 13;4:e06562. doi: 10.7554/eLife.06562. Elife. 2015. PMID: 25970034 Free PMC article. - The P286R mutation of DNA polymerase ε activates cancer-cell-intrinsic immunity and suppresses endometrial tumorigenesis via the cGAS-STING pathway.
Tang M, Yin S, Zeng H, Huang A, Huang Y, Hu Z, Shah AR, Zhang S, Li H, Chen G. Tang M, et al. Cell Death Dis. 2024 Jan 18;15(1):69. doi: 10.1038/s41419-023-06418-3. Cell Death Dis. 2024. PMID: 38238314 Free PMC article.
References
- Johnson A, O'Donnell M (2005) Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem 74: 283–315. - PubMed
- Johansson E, Macneill SA (2010) The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci 35: 339–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous