Chromatin as dynamic 10-nm fibers - PubMed (original) (raw)
Review
Chromatin as dynamic 10-nm fibers
Kazuhiro Maeshima et al. Chromosoma. 2014 Jun.
Abstract
Since Flemming described a nuclear substance in the nineteenth century and named it "chromatin," this substance has fascinated biologists. What is the structure of chromatin? DNA is wrapped around core histones, forming a nucleosome fiber (10-nm fiber). This fiber has long been assumed to fold into a 30-nm chromatin fiber and subsequently into helically folded larger fibers or radial loops. However, several recent studies, including our cryo-EM and X-ray scattering analyses, demonstrated that chromatin is composed of irregularly folded 10-nm fibers, without 30-nm chromatin fibers, in interphase chromatin and mitotic chromosomes. This irregular folding implies a chromatin state that is physically less constrained, which could be more dynamic compared with classical regular helical folding structures. Consistent with this, recently, we uncovered by single nucleosome imaging large nucleosome fluctuations in living mammalian cells (∼50 nm/30 ms). Subsequent computational modeling suggested that nucleosome fluctuation increases chromatin accessibility, which is advantageous for many "target searching" biological processes such as transcriptional regulation. Therefore, this review provides a novel view on chromatin structure in which chromatin consists of dynamic and disordered 10-nm fibers.
Figures
Fig. 1
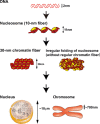
Old and novel views of chromatin structure. A long DNA molecule with a diameter of ∼2 nm is wrapped around a core histone octamer and forms a nucleosome with a diameter of 11 nm (Alberts et al. 2007). The nucleosome has long been assumed to fold into 30-nm chromatin fibers (left) and subsequently into the higher order organization of interphase nuclei or mitotic chromosomes. The right panel shows the novel hypothesis of irregularly folded nucleosome fibers
Fig. 2
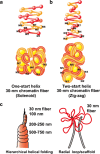
Two classical models of 30-nm chromatin fibers and higher order chromatin structures. a One-start helix (solenoid), b two-start helix (zigzag). (Top) A scheme of the two different topologies of chromatin fibers is shown (Robinson and Rhodes 2006). Positions from the first (N1) to the eighth (N8) nucleosome are labeled. c Two classical higher order chromatin structure models: the hierarchical helical folding model (Sedat and Manuelidis 1978) and the radial loop model (Laemmli et al. 1978). In the radial loop model, many loop structures of the 30-nm fiber (red) wrap around the scaffold structure (gray) (Laemmli et al. 1978), which consists of condensin and topoisomerase IIα (Maeshima and Laemmli 2003)
Fig. 3
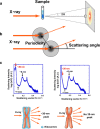
Small angle X-ray scattering (SAXS) analysis of chromatin structure. a Experimental design. The chromosome pellet in a quartz capillary tube was exposed to synchrotron X-ray beams, and the scattering patterns were recorded using the imaging plate (Nishino et al. 2012). b When non-crystal materials were irradiated with X-rays, scattering at small angles generally reflected periodic structures. Images a and b were reproduced from Joti et al. (2012), with some modifications. c Upper left Typical SAXS patterns of purified mitotic HeLa chromosome fractions. Three peaks at ∼6, ∼11 (weak), and ∼30 nm were detected (arrows). (Upper right) After the removal of ribosome aggregates, the 30-nm peak disappeared, whereas the other peaks remained. (Bottom) A model whereby the 30-nm peak in SAXS results from regularly spaced ribosome aggregates and not from the chromosomes. Image c was reproduced from Nishino et al. (2012), with some modification
Fig. 4
Polymer melt model. a Under low-salt conditions, nucleosome fibers could form 30-nm chromatin fibers via intra-fiber nucleosome associations. An increase in salt (cation) concentration results in inter-fiber nucleosomal contacts that interfere with intra-fiber nucleosomal associations, leading to a polymer melt scenario. Note that in these illustrations, we show a highly simplified two-dimensional nucleosome model. Arrows and dotted lines show repulsion forces and interactions, respectively. b During the melting process, the 30-nm chromatin fibers become irregularly folded nucleosome fibers
Fig. 5
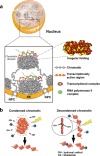
Higher order structure of interphase chromatin. a Condensed chromatin domains. Active chromatin regions are transcribed on the surfaces of chromatin domains with transcriptional complexes (purple spheres) and RNA polymerase II (green spheres). NPC nuclear pore complex, NE nuclear envelope. b (Left) Condensed chromatin is more resistant to radiation damage or chemical attack. (Right) Reactive radicals arising from the radiolysis of water molecules by irradiation can damage decondensed chromatin; decondensed chromatin is also more accessible to chemicals (labeled “Ch”)
Fig. 6
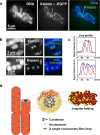
Mitotic chromosome structure. Axial localizations of condensins I and II in mitotic chromosomes in live mammalian cells. For DNA staining, DM (Indian Muntjac cells) cells stably expressing EGFP-Kleisin γ (condensin I) and EGFP-Kleisin β (condensing II) were stained with Hoechst 33342. Live-cell imaging was performed using a Delta Vision microscope (applied precision). a Clear axial signals of EGFP-Kleisin γ in mitotic chromosomes are shown. b End-on-view of mitotic chromosomes. The upper panel shows DM cells expressing EGFP-Kleisin γ, whereas the lower panel shows DM cells expressing EGFP-Kleisin β stably. Restricted dot signals from two types of EGFP-Kleisin in the cross-section of a chromosome body (DNA staining) are shown. c Quantitative data using line-profile analysis (blue line, DNA; red line, Kleisin signals) is shown. There is clear axial localization of condensins I and II in mitotic chromosomes in live mammalian cells. d Chromosomes consist of irregularly folded 10-nm nucleosome fibers. Condensins (blue) hold the nucleosome fibers (red) around the center of the chromosome. Locally, the nucleosome fibers are folded in an irregular or disordered manner, forming loop structures that collapse towards the center of the chromosome center (blue). The collapsed fiber (red) then forms a domain
Fig. 7
Single nucleosome imaging. a A small portion of PA-GFP-H4 was activated spontaneously without laser activation and was used for single nucleosome imaging. b Single nucleosome image of a DM cell (Indian Muntjac cell) nucleus that expresses PA-GFP-H4. PA-GFP-H4 is observed as a bright dot using oblique illumination microscopy. The dots were fitted to an assumed Gaussian point spread function to determine the precise center of signals with higher resolution. Bar = 5 μm. c Representative three trajectories of fluorescently tagged single nucleosomes. d Chromatin fluctuations as a basis for scanning genome information. In cells, nucleosome fibers (red spheres and lines) are folded irregularly. The nucleosomes fluctuate, and these nucleosome dynamics facilitate chromatin accessibility. The images were reproduced from (Hihara et al. ; Nozaki et al. 2013) with some modification
Similar articles
- Chromosomes without a 30-nm chromatin fiber.
Joti Y, Hikima T, Nishino Y, Kamada F, Hihara S, Takata H, Ishikawa T, Maeshima K. Joti Y, et al. Nucleus. 2012 Sep-Oct;3(5):404-10. doi: 10.4161/nucl.21222. Epub 2012 Jul 31. Nucleus. 2012. PMID: 22825571 Free PMC article. - Flexible and dynamic nucleosome fiber in living mammalian cells.
Nozaki T, Kaizu K, Pack CG, Tamura S, Tani T, Hihara S, Nagai T, Takahashi K, Maeshima K. Nozaki T, et al. Nucleus. 2013 Sep-Oct;4(5):349-56. doi: 10.4161/nucl.26053. Epub 2013 Aug 12. Nucleus. 2013. PMID: 23945462 Free PMC article. - Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure.
Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K. Nishino Y, et al. EMBO J. 2012 Apr 4;31(7):1644-53. doi: 10.1038/emboj.2012.35. Epub 2012 Feb 17. EMBO J. 2012. PMID: 22343941 Free PMC article. - Dynamic chromatin organization in the cell.
Prieto EI, Maeshima K. Prieto EI, et al. Essays Biochem. 2019 Apr 23;63(1):133-145. doi: 10.1042/EBC20180054. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 30967477 Review. - Structural transition of the nucleosome during chromatin remodeling and transcription.
Kobayashi W, Kurumizaka H. Kobayashi W, et al. Curr Opin Struct Biol. 2019 Dec;59:107-114. doi: 10.1016/j.sbi.2019.07.011. Epub 2019 Aug 29. Curr Opin Struct Biol. 2019. PMID: 31473439 Review.
Cited by
- The 3D Topography of Mitotic Chromosomes.
Chu L, Liang Z, Mukhina M, Fisher J, Vincenten N, Zhang Z, Hutchinson J, Zickler D, Kleckner N. Chu L, et al. Mol Cell. 2020 Sep 17;79(6):902-916.e6. doi: 10.1016/j.molcel.2020.07.002. Epub 2020 Aug 7. Mol Cell. 2020. PMID: 32768407 Free PMC article. - Coming to terms with chromatin structure.
Even-Faitelson L, Hassan-Zadeh V, Baghestani Z, Bazett-Jones DP. Even-Faitelson L, et al. Chromosoma. 2016 Mar;125(1):95-110. doi: 10.1007/s00412-015-0534-9. Epub 2015 Jul 30. Chromosoma. 2016. PMID: 26223534 Review. - Chromatin compaction under mixed salt conditions: opposite effects of sodium and potassium ions on nucleosome array folding.
Allahverdi A, Chen Q, Korolev N, Nordenskiöld L. Allahverdi A, et al. Sci Rep. 2015 Feb 17;5:8512. doi: 10.1038/srep08512. Sci Rep. 2015. PMID: 25688036 Free PMC article. - Contribution of advanced fluorescence nano microscopy towards revealing mitotic chromosome structure.
Botchway SW, Farooq S, Sajid A, Robinson IK, Yusuf M. Botchway SW, et al. Chromosome Res. 2021 Mar;29(1):19-36. doi: 10.1007/s10577-021-09654-5. Epub 2021 Mar 9. Chromosome Res. 2021. PMID: 33686484 Review. - Higher order genomic organization and epigenetic control maintain cellular identity and prevent breast cancer.
Fritz AJ, Gillis NE, Gerrard DL, Rodriguez PD, Hong D, Rose JT, Ghule PN, Bolf EL, Gordon JA, Tye CE, Boyd JR, Tracy KM, Nickerson JA, van Wijnen AJ, Imbalzano AN, Heath JL, Frietze SE, Zaidi SK, Carr FE, Lian JB, Stein JL, Stein GS. Fritz AJ, et al. Genes Chromosomes Cancer. 2019 Jul;58(7):484-499. doi: 10.1002/gcc.22731. Epub 2019 Mar 15. Genes Chromosomes Cancer. 2019. PMID: 30873710 Free PMC article. Review.
References
- Ahmed K, Li R, Bazett-Jones DP. Electron spectroscopic imaging of the nuclear landscape. Methods Mol Biol. 2009;464:415–423. - PubMed
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the Cell. 5. New York: Garland; 2007.
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, Yang S, Rohr K, Leonhardt H, Solovei I, Cremer C, Fakan S, Cremer T. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. - PubMed
- Asakura S, Oosawa F. On interaction between two bodies immersed in a solution of macromolecules. J Chem Phys. 1954;22:1255–1256.
- Bassett A, Cooper S, Wu C, Travers A. The folding and unfolding of eukaryotic chromatin. Curr Opin Genet Dev. 2009;19:159–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources