Implications of streamlining theory for microbial ecology - PubMed (original) (raw)
Review
Implications of streamlining theory for microbial ecology
Stephen J Giovannoni et al. ISME J. 2014 Aug.
Abstract
Whether a small cell, a small genome or a minimal set of chemical reactions with self-replicating properties, simplicity is beguiling. As Leonardo da Vinci reportedly said, 'simplicity is the ultimate sophistication'. Two diverging views of simplicity have emerged in accounts of symbiotic and commensal bacteria and cosmopolitan free-living bacteria with small genomes. The small genomes of obligate insect endosymbionts have been attributed to genetic drift caused by small effective population sizes (Ne). In contrast, streamlining theory attributes small cells and genomes to selection for efficient use of nutrients in populations where Ne is large and nutrients limit growth. Regardless of the cause of genome reduction, lost coding potential eventually dictates loss of function. Consequences of reductive evolution in streamlined organisms include atypical patterns of prototrophy and the absence of common regulatory systems, which have been linked to difficulty in culturing these cells. Recent evidence from metagenomics suggests that streamlining is commonplace, may broadly explain the phenomenon of the uncultured microbial majority, and might also explain the highly interdependent (connected) behavior of many microbial ecosystems. Streamlining theory is belied by the observation that many successful bacteria are large cells with complex genomes. To fully appreciate streamlining, we must look to the life histories and adaptive strategies of cells, which impose minimum requirements for complexity that vary with niche.
Figures
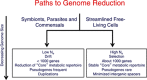
Figure 1
Alternate pathways of genome reduction. The mechanisms leading to small genomes in symbiotic and commensal organisms (for example, mycoplasmas) are fundamentally different from the process that lead to the small genomes that have recently been discovered in many free-living organisms from natural systems.
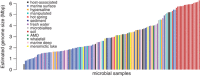
Figure 2
Average microbial genome sizes estimated from metagenomic data, adapted from Angly et al. (2009), with additional data from Frank and Sorensen (2011), Oh et al. (2011), Quaiser et al. (2011), Xia et al. (2011), Eiler et al. (2013).
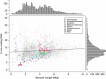
Figure 3
The % of noncoding (spacer) DNA versus genome length for all published bacterial genomes in IMG v400 and estimated genome size for single-amplified genomes (SAGs) from Swan et al. (2013). Streamlined (SAR11, Prochlorococcus, symbionts) and non-streamlined important marine taxa (Vibrionaceae, Rhodobacteraceae, Alteromonadaceae) have been highlighted (legend). Organisms where streamlining is driven by nutrient-limitation, such as SAR11 and those represented by the uncultured single-amplified genomes from Swan et al. (2013), have low % noncoding DNA and tend to fall at the extremes of the distribution of residuals from the linear regression line (black line). Conversely, organisms where streamlining is driven by symbiosis tend to maintain a broad range of % noncoding DNA, despite their small genomes. Histograms (top, right) show the distribution of points across each axis. The distribution of genome lengths (top) was significantly different to a unimodal distribution (Hartigan's Dip Test; Hartigan and Hartigan, 1985), _N_=5689, _D_=0.01, _P_=7.56 × 10−5) with a separation of the two modes at ∼3.7 Mbp.
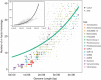
Figure 4
The number of σ-factor homologs versus genome length for published bacterial genomes in IMG v400 (grey), up to 5 Mbp in length with important marine microbial taxa highlighted (legend). Fitting the data across all genome lengths (inset) with a Poisson distribution showed evidence of overdispersion (Φ=5.50) therefore regression was performed using a negative binomial generalized linear model with a log-link (green line main figure, black line inset. Lighter green ribbon represents 95% confidence intervals of the model). The explained deviance of the negative binomial model was 0.73. Uncultured SAG representatives from Swan et al. (2013), SAR11, Prochlorococcus and symbionts fell below the regression line, supporting the hypothesis that low numbers of σ-factors are a consistent feature of streamlined genomes. SAGs from Verrucomicrobia fell above and below the regression line, supporting the findings of Swan et al. (2013) that these genomes can be found in both streamlined and non-streamlined varieties.
Similar articles
- Growth temperature and genome size in bacteria are negatively correlated, suggesting genomic streamlining during thermal adaptation.
Sabath N, Ferrada E, Barve A, Wagner A. Sabath N, et al. Genome Biol Evol. 2013;5(5):966-77. doi: 10.1093/gbe/evt050. Genome Biol Evol. 2013. PMID: 23563968 Free PMC article. - Streamlining and core genome conservation among highly divergent members of the SAR11 clade.
Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS. Grote J, et al. mBio. 2012 Sep 18;3(5):e00252-12. doi: 10.1128/mBio.00252-12. Print 2012. mBio. 2012. PMID: 22991429 Free PMC article. - Increased Mutation Rate Is Linked to Genome Reduction in Prokaryotes.
Bourguignon T, Kinjo Y, Villa-Martín P, Coleman NV, Tang Q, Arab DA, Wang Z, Tokuda G, Hongoh Y, Ohkuma M, Ho SYW, Pigolotti S, Lo N. Bourguignon T, et al. Curr Biol. 2020 Oct 5;30(19):3848-3855.e4. doi: 10.1016/j.cub.2020.07.034. Epub 2020 Aug 6. Curr Biol. 2020. PMID: 32763167 - Streamlining and simplification of microbial genome architecture.
Lynch M. Lynch M. Annu Rev Microbiol. 2006;60:327-49. doi: 10.1146/annurev.micro.60.080805.142300. Annu Rev Microbiol. 2006. PMID: 16824010 Review. - Endosymbiont evolution: predictions from theory and surprises from genomes.
Wernegreen JJ. Wernegreen JJ. Ann N Y Acad Sci. 2015 Dec;1360(1):16-35. doi: 10.1111/nyas.12740. Epub 2015 Apr 9. Ann N Y Acad Sci. 2015. PMID: 25866055 Free PMC article. Review.
Cited by
- Genome-resolved metagenomics identifies the particular genetic traits of phosphate-solubilizing bacteria in agricultural soil.
Wu X, Cui Z, Peng J, Zhang F, Liesack W. Wu X, et al. ISME Commun. 2022 Feb 16;2(1):17. doi: 10.1038/s43705-022-00100-z. ISME Commun. 2022. PMID: 37938650 Free PMC article. - Estuarine microbial networks and relationships vary between environmentally distinct communities.
Anderson SR, Harvey EL. Anderson SR, et al. PeerJ. 2022 Sep 20;10:e14005. doi: 10.7717/peerj.14005. eCollection 2022. PeerJ. 2022. PMID: 36157057 Free PMC article. - Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous 'streamer' community.
Beam JP, Jay ZJ, Schmid MC, Rusch DB, Romine MF, Jennings Rde M, Kozubal MA, Tringe SG, Wagner M, Inskeep WP. Beam JP, et al. ISME J. 2016 Jan;10(1):210-24. doi: 10.1038/ismej.2015.83. Epub 2015 Jul 3. ISME J. 2016. PMID: 26140529 Free PMC article. - Dispersing misconceptions and identifying opportunities for the use of 'omics' in soil microbial ecology.
Prosser JI. Prosser JI. Nat Rev Microbiol. 2015 Jul;13(7):439-46. doi: 10.1038/nrmicro3468. Epub 2015 Jun 8. Nat Rev Microbiol. 2015. PMID: 26052662 - Bacterioplankton seasonality in deep high-mountain lakes.
Zufiaurre A, Felip M, Camarero L, Sala-Faig M, Juhanson J, Bonilla-Rosso G, Hallin S, Catalan J. Zufiaurre A, et al. Front Microbiol. 2022 Sep 14;13:935378. doi: 10.3389/fmicb.2022.935378. eCollection 2022. Front Microbiol. 2022. PMID: 36187988 Free PMC article.
References
- Babu MM. (2003). Did the loss of sigma factors initiate pseudogene accumulation in M-leprae? Trends Microbiol 11: 59–61. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials