TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape - PubMed (original) (raw)
TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape
Koon Ho Wong et al. Mol Cell. 2014.
Abstract
The transition between transcriptional initiation and elongation by RNA polymerase (Pol) II is associated with phosphorylation of its C-terminal tail (CTD). Depletion of Kin28, the TFIIH subunit that phosphorylates the CTD, does not affect elongation but causes Pol II occupancy profiles to shift upstream in a FACT-independent manner indicative of a defect in promoter escape. Stronger defects in promoter escape are linked to stronger effects on preinitiation complex formation and transcription, suggesting that impairment in promoter escape results in premature dissociation of general factors and Pol II near the promoter. Kin28 has a stronger effect on genes whose transcription is dependent on SAGA as opposed to TFIID. Strikingly, Kin28 depletion causes a dramatic increase in Mediator at the core promoter. These observations suggest that TFIIH phosphorylation of the CTD causes Mediator dissociation, thereby permitting rapid promoter escape of Pol II from the preinitiation complex.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
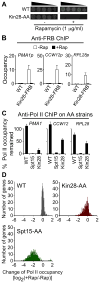
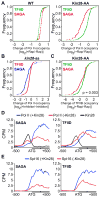
Figure 1. Kin28 is important, but not essential for transcription
(A) Growth spotting tests for wild-type (WT) and Kin28 anchor-away (AA) strains on rich YPD media with or without rapamycin at 30 °C for 2 days. (B) Kin28-FRB occupancy at the indicated promoters in the Kin28-AA and control strains before and after 1 hour of rapamycin treatment. Averages and standard errors of three individual experiments are shown. (C) Percentage of Pol II occupancy remained at the indicated coding regions in the WT, Spt15-AA and Kin28-AA strains after 1 hour of rapamycin treatment. Averages and standard errors of three individual experiments are presented. (D) Changes of Pol II occupancy (measured by Pol II ChIP-seq) at 3′ end of active genes in the Kin28-AA and Spt15-AA strains upon rapamycin treatment. Numbers of genes are plotted against the log2 ratios of Pol II levels after rapamycin treatment (+Rap) over Pol II levels before rapamycin treatment (−Rap). See also Figure S1.
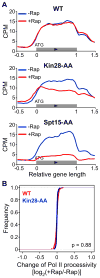
Figure 2. Kin28 depletion causes Pol II re-distribution to 5′ ends of genes, but does not affect Pol II processivity
(A) Overall Pol II ChIP-seq binding profiles of active genes in WT, Kin28-AA, and Spt15-AA strains before and after 1 hour of rapamycin treatment. (B) Changes of Pol II processivity (measured between +500–1000) of active genes greater than 1 kb in length in the indicated strains after 1 hour of rapamycin treatment. Changes of Pol II processivity are expressed as log2 ratios of gradients of the linear regression lines after and before rapamycin treatment. See also Figure S2.
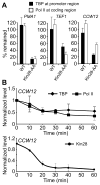
Figure 3. Kin28 depletion affects PIC formation
(A) TBP and Pol II occupancies at the promoter and the coding regions of three representative genes were analyzed before and after addition of rapamycin for 1 hour in the WT and Kin28-AA strains. Results are expressed as percentage of TBP and Pol II occupancies remained after the rapamycin treatment compared to those without the treatment. (B) Time course analysis of TBP, Pol II and Kin28-FRB occupancies at the promoter region of highly expressed CCW12 upon rapamycin treatment. Values shown are occupancy levels relative to those before rapamycin treatment (i.e. Time = 0 min). Averages and standard errors of three individual experiments are presented. See also Figure S3.
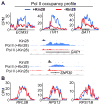
Figure 4. Kin28 depletion causes heterogeneous transcription effects
(A, B) Pol II ChIP-seq binding profiles and genome-browser display of the indicated genes in the Kin28-AA strain before (+Kin28) and after (−Kin28) 1 hour of rapamycin treatment. See also Figure S4.
Figure 5. Kin28 depletion affects SAGA-dependent transcription more than TFIID-dependent transcription
(A, B) Cumulative frequency plots of transcription (Pol II occupancy) changes at SAGA- and TFIID-dependent genes (Huisinga and Pugh, 2004) upon Kin28 depletion in the Kin28-AA strain or inactivation of Kin28 kinase activity in the _kin28_-as strain. (C) Cumulative frequency plots of TFIIB occupancy changes at SAGA- and TFIID-dependent genes upon Kin28 depletion by anchor-away. (D) Pol II (blue and red lines) and Kin28-FRB (black line) ChIP-seq binding profiles across promoter regions (+/− 500 bp from ATG) of active SAGA- and TFIID-dependent genes in the Kin28-AA strain before and after Kin28 depletion. (E) Spt16 ChIP-seq binding profiles were plotted for the same regions, as in (D). See also Figures S5 and S6.
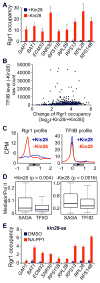
Figure 6. Kin28 depletion causes dramatic accumulation of Mediator at core promoters of all active genes
(A) Mediator (Rgr1) occupancy at the core promoters of indicated genes in the Kin28-AA strain before and after Kin28 depletion. Averages and standard errors of three individual experiments are presented. (B) Comparison between TFIIB levels and Mediator (Rgr1) binding changes in the Kin28-AA strain upon Kin28 depletion. Promoter TFIIB level was determined by the total count of TFIIB ChIP-seq reads within the 100 bp window centered on the transcription start site. For Mediator (Rgr1) binding changes, normalized read counts of analyzed promoters after Kin28 depletion were divided by those before Kin28 depletion and expressed as log2 ratios. Only promoters with more than 1000 TFIIB raw counts before Kin28 depletion were included in the analysis. (C) Mediator (Rgr1) and TFIIB ChIP-seq binding profiles of active genes in the Kin28-AA strain before (blue line) and after (red line) Kin28 depletion. (D) Boxplots of Mediator (Rgr1) to Pol II binding ratios at core promoter regions of active SAGA- and TFIID-dependent genes in the Kin28-AA strain before (+Kin28) and after (−Kin28) Kin28 depletion. (E) Mediator (Rgr1) occupancy at the core promoters of indicated genes in the _kin28_-as strain upon inactivation of Kin28 kinase activity by the inhibitor NA-PP1. Averages and standard errors of three individual experiments are presented. See also Figure S7.
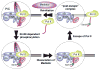
Figure 7. Model for the transition between initiation and elongation
The general transcription factors and Mediator assemble at a core promoter to form a pre-initiation complex (PIC). Upon PIC formation, Kin28-dependent phosphorylation of Pol II CTD inhibits the interaction between Pol II and Mediator, leading to a rapid dissociation of Mediator from the PIC, facilitating Pol II escape from the PIC for productive elongation. Although it is formally possible that Mediator dissociation and Pol II escape from the PIC occur simultaneously or in the reverse order, we disfavor this possibility as CTD phosphorylation destabilizes the direct Mediator-Pol II interaction in vitro, and there is no evidence in vivo for direct stable Mediator-PIC association in the absence of Pol II. After Pol II promoter escape, a “post-escape” transcription initiation complex, which is relatively stable (minute time scale), remains bound at the core promoter such that transcription re-initiation can occur upon reloading of Mediator and Pol II.
Similar articles
- Mutual targeting of mediator and the TFIIH kinase Kin28.
Guidi BW, Bjornsdottir G, Hopkins DC, Lacomis L, Erdjument-Bromage H, Tempst P, Myers LC. Guidi BW, et al. J Biol Chem. 2004 Jul 9;279(28):29114-20. doi: 10.1074/jbc.M404426200. Epub 2004 May 4. J Biol Chem. 2004. PMID: 15126497 - CDK7 kinase activity promotes RNA polymerase II promoter escape by facilitating initiation factor release.
Velychko T, Mohammad E, Ferrer-Vicens I, Parfentev I, Werner M, Studniarek C, Schwalb B, Urlaub H, Murphy S, Cramer P, Lidschreiber M. Velychko T, et al. Mol Cell. 2024 Jun 20;84(12):2287-2303.e10. doi: 10.1016/j.molcel.2024.05.007. Epub 2024 May 30. Mol Cell. 2024. PMID: 38821049 - Structures of the human Mediator and Mediator-bound preinitiation complex.
Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, Liu W, Xu Y. Chen X, et al. Science. 2021 Jun 4;372(6546):eabg0635. doi: 10.1126/science.abg0635. Epub 2021 May 6. Science. 2021. PMID: 33958484 - The Long Road to Understanding RNAPII Transcription Initiation and Related Syndromes.
Compe E, Egly JM. Compe E, et al. Annu Rev Biochem. 2021 Jun 20;90:193-219. doi: 10.1146/annurev-biochem-090220-112253. Annu Rev Biochem. 2021. PMID: 34153211 Review. - Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression.
Hirose Y, Ohkuma Y. Hirose Y, et al. J Biochem. 2007 May;141(5):601-8. doi: 10.1093/jb/mvm090. Epub 2007 Apr 3. J Biochem. 2007. PMID: 17405796 Review.
Cited by
- Architectural Mediator subunits are differentially essential for global transcription in Saccharomyces cerevisiae.
Tourigny JP, Schumacher K, Saleh MM, Devys D, Zentner GE. Tourigny JP, et al. Genetics. 2021 Mar 31;217(3):iyaa042. doi: 10.1093/genetics/iyaa042. Genetics. 2021. PMID: 33789343 Free PMC article. - The CTD Is Not Essential for the Post-Initiation Control of RNA Polymerase II Activity.
Gerber A, Roeder RG. Gerber A, et al. J Mol Biol. 2020 Sep 4;432(19):5489-5498. doi: 10.1016/j.jmb.2020.07.010. Epub 2020 Jul 21. J Mol Biol. 2020. PMID: 32707132 Free PMC article. - Simplicity is the Ultimate Sophistication-Crosstalk of Post-translational Modifications on the RNA Polymerase II.
Venkat Ramani MK, Yang W, Irani S, Zhang Y. Venkat Ramani MK, et al. J Mol Biol. 2021 Jul 9;433(14):166912. doi: 10.1016/j.jmb.2021.166912. Epub 2021 Mar 5. J Mol Biol. 2021. PMID: 33676925 Free PMC article. Review. - An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters.
Mittal C, Lang O, Lai WKM, Pugh BF. Mittal C, et al. Genes Dev. 2022 Sep 1;36(17-18):985-1001. doi: 10.1101/gad.350026.122. Epub 2022 Oct 27. Genes Dev. 2022. PMID: 36302553 Free PMC article. - A CUG codon-adapted anchor-away toolkit for functional analysis of genes in Candida albicans.
Teli BB, Nagar P, Priyadarshini Y, Poonia P, Natarajan K. Teli BB, et al. mSphere. 2024 Feb 28;9(2):e0070323. doi: 10.1128/msphere.00703-23. Epub 2024 Jan 22. mSphere. 2024. PMID: 38251906 Free PMC article.
References
- Akoulitchev S, Makela TP, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. - PubMed
- Bar-Joseph Z, Gifford DK, Jaakkola TS. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics. 2001;17:S22–29. - PubMed
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–170. - PubMed
- Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials