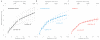Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas - PubMed (original) (raw)
Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas
Christopher J Russo et al. Nat Methods. 2014 Jun.
Abstract
Despite its many favorable properties as a sample support for biological electron microscopy, graphene is not widely used because its hydrophobicity precludes reliable protein deposition. We describe a method to modify graphene with a low-energy hydrogen plasma, which reduces hydrophobicity without degrading the graphene lattice. Use of plasma-treated graphene enables better control of protein distribution in ice for electron cryo-microscopy and improves image quality by reducing radiation-induced sample motion.
Figures
Figure 1. Low-energy hydrogen plasma treatment renders graphene hydrophilic and removes contamination.
(a) Plots of the air-water-graphene contact angle versus exposure time, with insets containing examples of optical micrographs used to measure the angles. The curve is an exponential fit to the data with rate constant of 1/58 seconds. Error bars are the std. err. at each plasma dose for the 3-5 measurements in y and the estimated accuracy of the exposure time, ± 1 second, in x. (b-d) Selected area electron diffraction patterns for the same suspended graphene sample: before hydrogen plasma exposure (b), after 20 seconds (c) and after 40 seconds (d). Arrow points to the 0-110 reflection at 2.14 Å and sets the scale for all three diffractograms. The change in the lattice constant for c & d relative to b is less than the error in the measurement, ≈0.9% (Supplementary Fig. 1). (e-f) Electron micrographs of suspended graphene before (e) and after (f) 30 second hydrogen plasma treatment respectively. Scale bars are 1000 Å, nominal defocus is −2.0 μm and fluence (electron dose) is 25 e−/Å2. (g) Power spectral density in each complete micrograph (Supplementary Fig. 2), normalized to the total image intensity, for 28 ± 1.4 Å thick am-C (Supplementary Fig. 2e), graphene before plasma treatment (e), and graphene after plasma treatment (f).
Figure 2. Dose dependent adsorption of proteins on hydrogen plasma-treated graphene.
(a) Electron cryo-micrographs of 70S ribosomes in vitrified ice at 80 K. Upper left quadrant is a standard grid treated with a 10 s hydrogen plasma dose. Other three quadrants show grids covered with monolayer graphene, and treated with 10, 20 and 40 s of hydrogen plasma. All other sample concentration, blotting, vitrification and imaging conditions are the same for all four grids. Scale bar is 1000 Å. (b-c) Electron micrographs of 20S proteasome (b) or apoferritin (c) molecules on graphene treated with 40 s of hydrogen plasma (lower panels) and molecules in unsupported ice from an adjacent region of the same grid (upper panels). Scale bars are 1000 Å and magnification is the same for (b) and (c).
Figure 3. Reduced motion of proteins on graphene substrates: speed plots.
The average 80S ribosome displacement from its initial position is plotted vs. time/ electron fluence for data collected in ice: supported by a continuous layer of am-C (a), without any support layer (b) and supported on a graphene substrate (c). Each point (dotted lines) represents the root mean squared (RMS) displacement of thousands of particles from a single grid (see Methods), whose positions were measured using a five-frame running average under constant electron beam irradiation (300 keV; 16 e−/Å2/s). Solid lines are the linear fits to the two phases of motion, with the slopes (ensemble particle speed) as indicated. Error bars are the standard error of the mean of the replicate experiments (3 separate grids for a,b 4 for c). All plots have the same scale.
Similar articles
- Improved sample dispersion in cryo-EM using "perpetually-hydrated" graphene oxide flakes.
Cheung M, Adaniya H, Cassidy C, Yamashita M, Li KL, Taba S, Shintake T. Cheung M, et al. J Struct Biol. 2018 Oct;204(1):75-79. doi: 10.1016/j.jsb.2018.07.008. Epub 2018 Jul 18. J Struct Biol. 2018. PMID: 30030043 - Graphene in cryo-EM specimen optimization.
Liu N, Wang HW. Liu N, et al. Curr Opin Struct Biol. 2024 Jun;86:102823. doi: 10.1016/j.sbi.2024.102823. Epub 2024 Apr 29. Curr Opin Struct Biol. 2024. PMID: 38688075 Review. - Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Zheng L, Liu N, Gao X, Zhu W, Liu K, Wu C, Yan R, Zhang J, Gao X, Yao Y, Deng B, Xu J, Lu Y, Liu Z, Li M, Wei X, Wang HW, Peng H. Zheng L, et al. Nat Methods. 2023 Jan;20(1):123-130. doi: 10.1038/s41592-022-01693-y. Epub 2022 Dec 15. Nat Methods. 2023. PMID: 36522503 Free PMC article. - A simple and robust procedure for preparing graphene-oxide cryo-EM grids.
Palovcak E, Wang F, Zheng SQ, Yu Z, Li S, Betegon M, Bulkley D, Agard DA, Cheng Y. Palovcak E, et al. J Struct Biol. 2018 Oct;204(1):80-84. doi: 10.1016/j.jsb.2018.07.007. Epub 2018 Jul 11. J Struct Biol. 2018. PMID: 30017701 Free PMC article. - Application of Monolayer Graphene and Its Derivative in Cryo-EM Sample Preparation.
Wu K, Wu D, Zhu L, Wu Y. Wu K, et al. Int J Mol Sci. 2021 Aug 19;22(16):8940. doi: 10.3390/ijms22168940. Int J Mol Sci. 2021. PMID: 34445650 Free PMC article. Review.
Cited by
- Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity.
Nogales E, Scheres SH. Nogales E, et al. Mol Cell. 2015 May 21;58(4):677-89. doi: 10.1016/j.molcel.2015.02.019. Mol Cell. 2015. PMID: 26000851 Free PMC article. Review. - Cryo-electron tomography related radiation-damage parameters for individual-molecule 3D structure determination.
Xue H, Zhang M, Liu J, Wang J, Ren G. Xue H, et al. Front Chem. 2022 Aug 30;10:889203. doi: 10.3389/fchem.2022.889203. eCollection 2022. Front Chem. 2022. PMID: 36110139 Free PMC article. Review. - Biological Applications at the Cutting Edge of Cryo-Electron Microscopy.
Dillard RS, Hampton CM, Strauss JD, Ke Z, Altomara D, Guerrero-Ferreira RC, Kiss G, Wright ER. Dillard RS, et al. Microsc Microanal. 2018 Aug;24(4):406-419. doi: 10.1017/S1431927618012382. Microsc Microanal. 2018. PMID: 30175702 Free PMC article. Review. - Cryo-EM Grid Preparation of Membrane Protein Samples for Single Particle Analysis.
Sgro GG, Costa TRD. Sgro GG, et al. Front Mol Biosci. 2018 Jul 31;5:74. doi: 10.3389/fmolb.2018.00074. eCollection 2018. Front Mol Biosci. 2018. PMID: 30131964 Free PMC article. Review. - Multifunctional graphene supports for electron cryomicroscopy.
Naydenova K, Peet MJ, Russo CJ. Naydenova K, et al. Proc Natl Acad Sci U S A. 2019 Jun 11;116(24):11718-11724. doi: 10.1073/pnas.1904766116. Epub 2019 May 24. Proc Natl Acad Sci U S A. 2019. PMID: 31127045 Free PMC article.
References
- Dubochet J, et al. Q Rev Biophys. 1988;21:129–228. - PubMed
- Robertson J. Advances in Physics. 1986;35:317–374.
- Miyazawa A, et al. J Mol Biol. 1999;288:765–786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources