Comparison of methods for fecal microbiome biospecimen collection - PubMed (original) (raw)
Comparison of methods for fecal microbiome biospecimen collection
Christine Dominianni et al. BMC Microbiol. 2014.
Abstract
Background: Effective means are needed to efficiently collect fecal samples for microbiome analysis in large-scale epidemiological studies. Using twenty-four fecal aliquots prepared from three healthy individuals, we compared the following four fecal sample collection methods for assessment of human gut microbiome: 1) fecal occult blood test cards, held at room temperature for three days, 2) Eppendorf tubes, at room temperature for three days, 3) Eppendorf tubes with RNAlater, at room temperature, and 4) as controls, samples immediately frozen at -80°C. The 24 samples were assayed by 16S rRNA gene sequencing to compare overall microbiome structure and taxon distributions according to collection method.
Results: Storing fecal occult blood test card samples at room temperature for three days did not affect total DNA purity and relative 16S rRNA bacterial gene contents, compared with fresh frozen collection. Overall microbiome structure, based on phylogenetic UniFrac index, differed significantly by subject (p = 0.001), but microbiome structure (p = 0.497) and relative abundance of major microbial taxa (phyla) (p > 0.05) did not differ significantly by collection method.
Conclusions: Our findings suggest that low-cost fecal occult blood test card collection may be a feasible means of sample collection for fecal microbiome assessment in large-scale population-based studies.
Figures
Figure 1
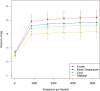
Alpha rarefaction plot of Shannon indices (±Standard Error) according to collection method. Card (green), Room Temperature (blue), RNAlater (orange), Frozen (red). Statistical significance was tested by using non-parametric Monte Carlo permutations (QIIME).
Figure 2
Unweighted PCoA plots of the first two principal coordinates. A), B) The first two principal coordinates were grouped by subject (1 [red], 2 [blue], 3 [orange]) A) or collection method (card [green], room temperature [blue], RNAlater [orange], frozen [red]) B). Adonis was used to test for significant differences in the variation in distances across subjects or collection methods using QIIME. C) UPGMA clustering on unweighted UniFrac distances (subject 1 [red], 2 [blue], 3 [orange]). D) Mean (±Std) unweighted UniFrac distances within and between sample collection methods or subjects.
Figure 3
Relative abundances of phyla by subject and by collection method. Card (1A-3A), Room Temperature (1B-3B), RNAlater (1C-3C), Frozen (1D-3D). Kruskal-Wallis or Mann-Whitney-Wilcoxon tests were used to test for overall differences using SAS software (version 9.3).
Similar articles
- Comparison of Fecal Collection Methods for Microbiome and Metabolomics Studies.
Wang Z, Zolnik CP, Qiu Y, Usyk M, Wang T, Strickler HD, Isasi CR, Kaplan RC, Kurland IJ, Qi Q, Burk RD. Wang Z, et al. Front Cell Infect Microbiol. 2018 Aug 28;8:301. doi: 10.3389/fcimb.2018.00301. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30234027 Free PMC article. - Comparison of Methods To Collect Fecal Samples for Microbiome Studies Using Whole-Genome Shotgun Metagenomic Sequencing.
Byrd DA, Sinha R, Hoffman KL, Chen J, Hua X, Shi J, Chia N, Petrosino J, Vogtmann E. Byrd DA, et al. mSphere. 2020 Feb 26;5(1):e00827-19. doi: 10.1128/mSphere.00827-19. mSphere. 2020. PMID: 32250964 Free PMC article. - Collection of non-meconium stool on fecal occult blood cards is an effective method for fecal microbiota studies in infants.
Wong WSW, Clemency N, Klein E, Provenzano M, Iyer R, Niederhuber JE, Hourigan SK. Wong WSW, et al. Microbiome. 2017 Sep 5;5(1):114. doi: 10.1186/s40168-017-0333-z. Microbiome. 2017. PMID: 28870234 Free PMC article. - Effects of Stool Sample Preservation Methods on Gut Microbiota Biodiversity: New Original Data and Systematic Review with Meta-Analysis.
Li XM, Shi X, Yao Y, Shen YC, Wu XL, Cai T, Liang LX, Wang F. Li XM, et al. Microbiol Spectr. 2023 Jun 15;11(3):e0429722. doi: 10.1128/spectrum.04297-22. Epub 2023 Apr 24. Microbiol Spectr. 2023. PMID: 37093040 Free PMC article. - Current Trends and Challenges of Microbiome Research in Prostate Cancer.
Trecarten S, Fongang B, Liss M. Trecarten S, et al. Curr Oncol Rep. 2024 May;26(5):477-487. doi: 10.1007/s11912-024-01520-x. Epub 2024 Apr 4. Curr Oncol Rep. 2024. PMID: 38573440 Review.
Cited by
- Methods for exploring the faecal microbiome of premature infants: a review.
Westaway JAF, Huerlimann R, Miller CM, Kandasamy Y, Norton R, Rudd D. Westaway JAF, et al. Matern Health Neonatol Perinatol. 2021 Mar 8;7(1):11. doi: 10.1186/s40748-021-00131-9. Matern Health Neonatol Perinatol. 2021. PMID: 33685524 Free PMC article. Review. - Systematic Analysis of Impact of Sampling Regions and Storage Methods on Fecal Gut Microbiome and Metabolome Profiles.
Liang Y, Dong T, Chen M, He L, Wang T, Liu X, Chang H, Mao JH, Hang B, Snijders AM, Xia Y. Liang Y, et al. mSphere. 2020 Jan 8;5(1):e00763-19. doi: 10.1128/mSphere.00763-19. mSphere. 2020. PMID: 31915218 Free PMC article. - Comparison of Fecal Collection Methods for Microbiome and Metabolomics Studies.
Wang Z, Zolnik CP, Qiu Y, Usyk M, Wang T, Strickler HD, Isasi CR, Kaplan RC, Kurland IJ, Qi Q, Burk RD. Wang Z, et al. Front Cell Infect Microbiol. 2018 Aug 28;8:301. doi: 10.3389/fcimb.2018.00301. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30234027 Free PMC article. - The standardisation of the approach to metagenomic human gut analysis: from sample collection to microbiome profiling.
Szóstak N, Szymanek A, Havránek J, Tomela K, Rakoczy M, Samelak-Czajka A, Schmidt M, Figlerowicz M, Majta J, Milanowska-Zabel K, Handschuh L, Philips A. Szóstak N, et al. Sci Rep. 2022 May 19;12(1):8470. doi: 10.1038/s41598-022-12037-3. Sci Rep. 2022. PMID: 35589762 Free PMC article. - Home-Made Cost Effective Preservation Buffer Is a Better Alternative to Commercial Preservation Methods for Microbiome Research.
Menke S, Gillingham MA, Wilhelm K, Sommer S. Menke S, et al. Front Microbiol. 2017 Jan 31;8:102. doi: 10.3389/fmicb.2017.00102. eCollection 2017. Front Microbiol. 2017. PMID: 28197142 Free PMC article.
References
- A guide to bacteria preservation: refrigeration, freezing and freeze drying. OPS Diagnositics. Accessed 10 March 2014, http://www.opsdiagnostics.com/notes/ranpri/aguidetobacteriapreservation.htm.
- Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Li NLH, Bushman FD. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. - DOI - PMC - PubMed
- Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdar AP, Kato I. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods. 2008;72(2):124–132. doi: 10.1016/j.mimet.2007.11.007. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical