Innate immunity to influenza virus infection - PubMed (original) (raw)
Review
Innate immunity to influenza virus infection
Akiko Iwasaki et al. Nat Rev Immunol. 2014 May.
Abstract
Influenza viruses are a major pathogen of both humans and animals. Recent studies using gene-knockout mice have led to an in-depth understanding of the innate sensors that detect influenza virus infection in a variety of cell types. Signalling downstream of these sensors induces distinct sets of effector mechanisms that block virus replication and promote viral clearance by inducing innate and adaptive immune responses. In this Review, we discuss the various ways in which the innate immune system uses pattern recognition receptors to detect and respond to influenza virus infection. We consider whether the outcome of innate sensor stimulation promotes antiviral resistance or disease tolerance, and propose rational treatment strategies for the acute respiratory disease that is caused by influenza virus infection.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
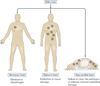
Figure 1. Antiviral resistance and disease tolerance
There are two distinct host strategies that deal with an infection: elimination of the pathogen or reduction of the negative impact of infection. A resistant host regains fitness by recognizing and eliminating the pathogen. A tolerant host regains fitness by reducing the immunopathology and tissue damage that is inflicted by pathogens. A host becomes susceptible if they are unable to reduce pathogen burden or unable to tolerate the negative consequences of the immune response to infection.
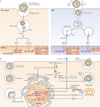
Figure 2. Innate sensing of influenza virus infection
Influenza virus infection is detected by multiple host sensors that recognize unique features that are associated with the infection. a | Infected cells are phagocytosed by macrophages (left panel) for recognition of double-stranded RNA (dsRNA) by Toll-like receptor 3 (TLR3), which leads to the induction of the expression of nuclear factor-κB (NF-κB)-dependent pro-inflammatory cytokines and of type I interferon (IFN) and IFN-stimulated genes (ISGs) downstream of IFN-regulatory factor 3 (IRF3). Incoming genomic single-stranded RNA (ssRNA) that is contained within the virion is released via the degradation of the viral membrane and capsid within acidified endosomes, and ssRNA is recognized by TLR7 in plasmacytoid dendritic cells (pDCs; right panel). TLR7 signalling induces NF-κB-dependent genes from the NF-κB endosome, and IRF7 activation from the IRF7 endosome. b | Within infected cells, viral RNA in the cytosol (possibly in stress granules; not shown) is recognized by retinoic acid-inducible gene I (RIG-I), which, through the activation of mitochondrial antiviral signalling protein(MAVS), leads to the induction of pro-inflammatory cytokines and type I IFN. Matrix 2 (M2) ion channel activity in the Golgi stimulates formation of the NOD-, LRR- and pyrin domain-containing 3 (NLRP3) inflammasome, which results in caspase 1 activation and the release of the cytokines interleukin-1β (IL-1β) and IL-18. PB1-F2 fibrils accumulate in the phagosome, which results in the activation of NLRP3 and the release of IL-1β and IL-18. ER, endoplasmic reticulum; HA, haemagglutinin; NA, neuraminidase; TRIF, TIR-domain-containing adaptor protein inducing IFNβ.
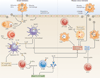
Figure 3. Mechanisms of resistance, disease and tolerance
Innate resistance (left) is conferred by type I and type III interferons (IFNs) that are secreted upon stimulation of retinoic acid-inducible gene I (RIG-I) in infected cells and Toll-like receptor 7 (TLR7) in plasmacytoid dendritic cells (pDCs). Type I IFNs act on most cells, whereas type III IFN (IFNλ) acts on epithelial cells to block virus replication. DCs and macrophages that are infected with influenza virus release interleukin-1β (IL-1β), which enables bystander DCs to upregulate CC-chemokine receptor 7 (CCR7) expression and migrate to the draining lymph nodes to stimulate T cells. NLRP3 inflammasomes also increase disease tolerance by promoting tissue repair. T cells and natural killer (NK) cells secrete IFNγ to induce an antiviral state or induce the granzyme B-mediated lysis of virus-infected cells, whereas B cells secrete antibodies to viral antigens to mediate adaptive immune protection of the host. Inflammatory cytokines (such as IL-1, tumour necrosis factor (TNF) and IL-17) that are induced as a result of innate signalling can also lead to pathology when the duration and extent of cytokine release is increased. Negative regulators of cytokines and inflammatory cells — such as IL-10 and CD200-CD200R — suppress inflammatory consequences, while positive regulators of tissue repair — such as amphiregulin and transforming growth factor-β (TGFβ) — promote a return to homeostasis. The balance between these negative and positive regulators determines whether the host succumbs to disease or can enter a state of tolerance (right). ILC2, type 2 innate lymphoid cell; NLRP3, NOD-, LRR- and pyrin domain-containing 3; ssRNA, single-stranded RNA.
Similar articles
- Innate Immune Responses to Influenza Virus Infections in the Upper Respiratory Tract.
Mifsud EJ, Kuba M, Barr IG. Mifsud EJ, et al. Viruses. 2021 Oct 17;13(10):2090. doi: 10.3390/v13102090. Viruses. 2021. PMID: 34696520 Free PMC article. Review. - Innate Immunity and the Inter-exposure Interval Determine the Dynamics of Secondary Influenza Virus Infection and Explain Observed Viral Hierarchies.
Cao P, Yan AW, Heffernan JM, Petrie S, Moss RG, Carolan LA, Guarnaccia TA, Kelso A, Barr IG, McVernon J, Laurie KL, McCaw JM. Cao P, et al. PLoS Comput Biol. 2015 Aug 18;11(8):e1004334. doi: 10.1371/journal.pcbi.1004334. eCollection 2015 Aug. PLoS Comput Biol. 2015. PMID: 26284917 Free PMC article. - Influenza Virus RNA Synthesis and the Innate Immune Response.
Weis S, Te Velthuis AJW. Weis S, et al. Viruses. 2021 Apr 28;13(5):780. doi: 10.3390/v13050780. Viruses. 2021. PMID: 33924859 Free PMC article. Review. - Inflammasome recognition of influenza virus is essential for adaptive immune responses.
Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Ichinohe T, et al. J Exp Med. 2009 Jan 16;206(1):79-87. doi: 10.1084/jem.20081667. Epub 2009 Jan 12. J Exp Med. 2009. PMID: 19139171 Free PMC article. - Role of γδ T cells in controlling viral infections with a focus on influenza virus: implications for designing novel therapeutic approaches.
Sabbaghi A, Miri SM, Keshavarz M, Mahooti M, Zebardast A, Ghaemi A. Sabbaghi A, et al. Virol J. 2020 Nov 12;17(1):174. doi: 10.1186/s12985-020-01449-0. Virol J. 2020. PMID: 33183352 Free PMC article. Review.
Cited by
- Downregulation of BmSTAT transcription factor promoted nucleopolyhedrovirus replication in Bombyx mori.
Liang W, Zhou L, Dang Z, Wang S, Zhao P, Xia Q, Lu Z. Liang W, et al. Front Microbiol. 2024 Oct 23;15:1485951. doi: 10.3389/fmicb.2024.1485951. eCollection 2024. Front Microbiol. 2024. PMID: 39507331 Free PMC article. - Fei-Yan-Qing-Hua decoction attenuates influenza virus infection by enhancing host antiviral response through microbiota-derived acetate.
Dou B, Wu X, He Y, Xu G, Zhang H, Huang Q, Chen X, Duan N, Zhou L, Zhang W, An H, Zheng Y. Dou B, et al. Front Pharmacol. 2024 Oct 10;15:1446749. doi: 10.3389/fphar.2024.1446749. eCollection 2024. Front Pharmacol. 2024. PMID: 39449967 Free PMC article. - Host combats porcine reproductive and respiratory syndrome virus infection at non-coding RNAs level.
Qin Z, Liu W, Qin Z, Zhang H, Huang X. Qin Z, et al. Virulence. 2024 Dec;15(1):2416551. doi: 10.1080/21505594.2024.2416551. Epub 2024 Oct 18. Virulence. 2024. PMID: 39403796 Free PMC article. Review. - Multifaceted roles of mitochondria in asthma.
Zhang W, Zhang C, Zhang Y, Zhou X, Dong B, Tan H, Su H, Sun X. Zhang W, et al. Cell Biol Toxicol. 2024 Oct 9;40(1):85. doi: 10.1007/s10565-024-09928-8. Cell Biol Toxicol. 2024. PMID: 39382744 Free PMC article. Review. - A clinical protocol for a German birth cohort study of the Maturation of Immunity Against respiratory viral Infections (MIAI).
Hartmann CR, Khan R, Schöning J, Richter M, Willers M, Pirr S, Heckmann J, Dirks J, Morbach H, Konrad M, Fries E, Winkler M, Büchel J, Seidenspinner S, Fischer J, Vollmuth C, Meinhardt M, Marissen J, Schmolke M, Haid S, Pietschmann T, Backes S, Dölken L, Löber U, Keil T, Heuschmann PU, Wöckel A, Sagar, Ulas T, Forslund-Startceva SK, Härtel C, Viemann D. Hartmann CR, et al. Front Immunol. 2024 Sep 17;15:1443665. doi: 10.3389/fimmu.2024.1443665. eCollection 2024. Front Immunol. 2024. PMID: 39355253 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- AI064705/AI/NIAID NIH HHS/United States
- AI054359/AI/NIAID NIH HHS/United States
- R01 AI064705/AI/NIAID NIH HHS/United States
- R01 AI081884/AI/NIAID NIH HHS/United States
- R01 AI054359/AI/NIAID NIH HHS/United States
- AI062428/AI/NIAID NIH HHS/United States
- AI081884/AI/NIAID NIH HHS/United States
- U54AI057160/AI/NIAID NIH HHS/United States
- R56 AI062428/AI/NIAID NIH HHS/United States
- R01 AI062428/AI/NIAID NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources