Holliday junction resolution: regulation in space and time - PubMed (original) (raw)
Review
Holliday junction resolution: regulation in space and time
Joao Matos et al. DNA Repair (Amst). 2014 Jul.
Abstract
Holliday junctions (HJs) can be formed between sister chromatids or homologous chromosomes during the recombinational repair of DNA lesions. A variety of pathways act upon HJs to remove them from DNA, in events that are critical for appropriate chromosome segregation. Despite the identification and characterization of multiple enzymes involved in HJ processing, the cellular mechanisms that regulate and implement pathway usage have only just started to be delineated. A conserved network of core cell-cycle kinases and phosphatases modulate HJ metabolism by exerting spatial and temporal control over the activities of two structure-selective nucleases: yeast Mus81-Mms4 (human MUS81-EME1) and Yen1 (human GEN1). These regulatory cycles operate to establish the sequential activation of HJ processing enzymes, implementing a hierarchy in pathway usage that ensure the elimination of chromosomal interactions which would otherwise interfere with chromosome segregation. Mus81-Mms4/EME1 and Yen1/GEN1 emerge to define a special class of enzymes, evolved to satisfy the cellular need of safeguarding the completion of DNA repair when on the verge of chromosome segregation.
Keywords: Cdc5; Cell-cycle; DNA repair; EME1; GEN1; Mms4; Mus81; Nuclease; PLK1; Recombination; Resolvase; Slx1; Slx4; Yen1.
Copyright © 2014 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
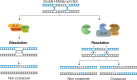
Fig. 1
DNA centric view showing three distinct pathways of Holliday junction processing. The BTR complex disengages dHJs, using the mechanism of “dissolution”, to generate NCO recombinants. MUS81-EME1 and SLX1-SLX4 interact to form the SLX-MUS complex which resolves both single and double HJs by endonucleolytic cleavage to generate COs and NCOs. GEN1 provides a separate pathway of HJ resolution.
Fig. 2
Cell-cycle centric view of HJ processing. During DNA repair, single or double HJs (sHJ or dHJ) are formed between sister chromatids or homologous chromosomes (for convenience, the diagram depicts two homologs). The STR (yeast) or BTR (human) complexes operate early during the cell cycle to dissolve dHJs and generate non-crossover recombinants. If DNA damage occurs late during the cell cycle or if HJs escape the attention of STR/BTR and persist until S/G2 phase, resolution enzymes ensure HJ processing and safeguard chromosome segregation.
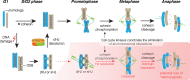
Fig. 3
Spatio-temporal regulation of HJ processing enzymes. In S. cerevisiae, cell cycle kinases Cdk and Cdc5 transiently phosphorylate and activate Mus81-Mms4 at the G2/M transition. Cdk1-mediated phosphorylation of Yen1 prevents its nuclear accumulation and biochemical activation during S-phase and early stages of mitosis. Cyclin degradation and Cdc14-mediated dephosphorylation activate Yen1 during anaphase. In human cells, a similar regulatory network uses a different molecular mechanism to prevent the actions of MUS81-EME1 and GEN1 during S-phase and early G2. At the G2/M transition, M-phase CDK activity promotes the interaction of MUS81-EME1 with SLX1-SLX4 leading to the formation of an active SLX-MUS HJ resolvase. GEN1, which is excluded from the nucleus during early stages of the cell cycle, gains access to chromatin upon CDK1-mediated nuclear envelope breakdown.
Similar articles
- Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions.
Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A. Garner E, et al. Cell Rep. 2013 Oct 17;5(1):207-15. doi: 10.1016/j.celrep.2013.08.041. Epub 2013 Sep 27. Cell Rep. 2013. PMID: 24080495 Free PMC article. - Mus81-Mms4 and Yen1 resolve a novel anaphase bridge formed by noncanonical Holliday junctions.
García-Luis J, Machín F. García-Luis J, et al. Nat Commun. 2014 Dec 3;5:5652. doi: 10.1038/ncomms6652. Nat Commun. 2014. PMID: 25466415 - Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells.
Wyatt HD, Sarbajna S, Matos J, West SC. Wyatt HD, et al. Mol Cell. 2013 Oct 24;52(2):234-47. doi: 10.1016/j.molcel.2013.08.035. Epub 2013 Sep 26. Mol Cell. 2013. PMID: 24076221 - Holliday junction processing enzymes as guardians of genome stability.
Sarbajna S, West SC. Sarbajna S, et al. Trends Biochem Sci. 2014 Sep;39(9):409-19. doi: 10.1016/j.tibs.2014.07.003. Epub 2014 Aug 14. Trends Biochem Sci. 2014. PMID: 25131815 Review. - Resolution of Recombination Intermediates: Mechanisms and Regulation.
West SC, Blanco MG, Chan YW, Matos J, Sarbajna S, Wyatt HD. West SC, et al. Cold Spring Harb Symp Quant Biol. 2015;80:103-9. doi: 10.1101/sqb.2015.80.027649. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370409 Review.
Cited by
- Checkpoints are blind to replication restart and recombination intermediates that result in gross chromosomal rearrangements.
Mohebi S, Mizuno K, Watson A, Carr AM, Murray JM. Mohebi S, et al. Nat Commun. 2015 Feb 27;6:6357. doi: 10.1038/ncomms7357. Nat Commun. 2015. PMID: 25721418 Free PMC article. - Homologous recombination maintenance of genome integrity during DNA damage tolerance.
Prado F. Prado F. Mol Cell Oncol. 2014 Oct 29;1(2):e957039. doi: 10.4161/23723548.2014.957039. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308329 Free PMC article. Review. - Biochemical attributes of mitotic and meiotic presynaptic complexes.
Crickard JB, Greene EC. Crickard JB, et al. DNA Repair (Amst). 2018 Nov;71:148-157. doi: 10.1016/j.dnarep.2018.08.018. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30195641 Free PMC article. Review. - A cytological revisit on parthenogenetic Artemia and the deficiency of a meiosis-specific recombinase DMC1 in the possible transition from bisexuality to parthenogenesis.
Xu LY, Wu WT, Bi N, Yan ZJ, Yang F, Yang WJ, Yang JS. Xu LY, et al. Chromosoma. 2023 Jun;132(2):89-103. doi: 10.1007/s00412-023-00790-x. Epub 2023 Mar 20. Chromosoma. 2023. PMID: 36939898 - The Slx4-Dpb11 scaffold complex: coordinating the response to replication fork stalling in S-phase and the subsequent mitosis.
Princz LN, Gritenaite D, Pfander B. Princz LN, et al. Cell Cycle. 2015;14(4):488-94. doi: 10.4161/15384101.2014.989126. Cell Cycle. 2015. PMID: 25496009 Free PMC article.
References
- Holliday R. A mechanism for gene conversion in fungi. Genet. Res. Camb. 1964;5:282–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous