CRISPR-Cas systems: new players in gene regulation and bacterial physiology - PubMed (original) (raw)
Review
CRISPR-Cas systems: new players in gene regulation and bacterial physiology
Timothy R Sampson et al. Front Cell Infect Microbiol. 2014.
Abstract
CRISPR-Cas systems are bacterial defenses against foreign nucleic acids derived from bacteriophages, plasmids or other sources. These systems are targeted in an RNA-dependent, sequence-specific manner, and are also adaptive, providing protection against previously encountered foreign elements. In addition to their canonical function in defense against foreign nucleic acid, their roles in various aspects of bacterial physiology are now being uncovered. We recently revealed a role for a Cas9-based Type II CRISPR-Cas system in the control of endogenous gene expression, a novel form of prokaryotic gene regulation. Cas9 functions in association with two small RNAs to target and alter the stability of an endogenous transcript encoding a bacterial lipoprotein (BLP). Since BLPs are recognized by the host innate immune protein Toll-like Receptor 2 (TLR2), CRISPR-Cas-mediated repression of BLP expression facilitates evasion of TLR2 by the intracellular bacterial pathogen Francisella novicida, and is essential for its virulence. Here we describe the Cas9 regulatory system in detail, as well as data on its role in controlling virulence traits of Neisseria meningitidis and Campylobacter jejuni. We also discuss potential roles of CRISPR-Cas systems in the response to envelope stress and other aspects of bacterial physiology. Since ~45% of bacteria and ~83% of Archaea encode these machineries, the newly appreciated regulatory functions of CRISPR-Cas systems are likely to play broad roles in controlling the pathogenesis and physiology of diverse prokaryotes.
Keywords: CRISPR-Cas; Cas9; Francisella novicida; bacterial pathogenesis; post-transcriptional regulation of gene expression.
Figures
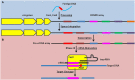
Figure 1
Function of the Type II CRISPR-Cas system in adaptive nucleic acid restriction. (A) Foreign DNA is recognized by Cas1 and Cas2 and is processed into a new spacer sequence (red) within the CRISPR array (Adaptation phase, blue). (B) To restrict foreign DNA, the CRISPR array is transcribed as a single transcript (pre-crRNA array) and matured into small targeting crRNAs in a process requiring RNase III and tracrRNA. The dsRNA complex of crRNA and tracrRNA is associated with Cas9 and the spacer sequence within the crRNA can hybridize to complementary DNA sequences. Cas9 then mediates cleavage of the targeted DNA downstream of the proto-spacer adjacent motif, or PAM, highlighted by the red circle (Effector phase, pink).
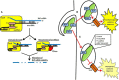
Figure 2
CRISPR-Cas-mediated gene regulation and innate immune evasion by F. novicida. (A) A dual-RNA complex consisting of the tracrRNA and scaRNA forms through interaction of a sequence identical to the CRISPR repeat (pink box). This dsRNA structure is associated with F. novicida Cas9 and allows the free portion of the tracrRNA to interact through a non-identity interaction with mRNA encoding the BLP (FTN_1103; blue). Subsequently, the stability of the BLP mRNA is altered, possibly via catalytic activity of Cas9 (cleavage event indicated by red triangle) or by an unknown RNase (red sector). (B) Following entry into host cells, TLR2 is capable of detecting BLPs present in the F. novicida envelope. This leads to activation of a proinflammatory cytokine response. (C) However, by upregulating CRISPR-Cas components after entry and while in the phagosome, F. novicida limits its BLP content and dampens the activation of TLR2, leading to decreased proinflammatory cytokine signaling.
Similar articles
- Degeneration of a CRISPR/Cas system and its regulatory target during the evolution of a pathogen.
Sampson TR, Weiss DS. Sampson TR, et al. RNA Biol. 2013 Oct;10(10):1618-22. doi: 10.4161/rna.26423. Epub 2013 Sep 20. RNA Biol. 2013. PMID: 24100224 Free PMC article. - Francisella novicida CRISPR-Cas Systems Can Functionally Complement Each Other in DNA Defense while Providing Target Flexibility.
Ratner HK, Weiss DS. Ratner HK, et al. J Bacteriol. 2020 May 27;202(12):e00670-19. doi: 10.1128/JB.00670-19. Print 2020 May 27. J Bacteriol. 2020. PMID: 32284320 Free PMC article. - Cas9-dependent endogenous gene regulation is required for bacterial virulence.
Sampson TR, Weiss DS. Sampson TR, et al. Biochem Soc Trans. 2013 Dec;41(6):1407-11. doi: 10.1042/BST20130163. Biochem Soc Trans. 2013. PMID: 24256228 Review. - A CRISPR/Cas system mediates bacterial innate immune evasion and virulence.
Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. Sampson TR, et al. Nature. 2013 May 9;497(7448):254-7. doi: 10.1038/nature12048. Epub 2013 Apr 14. Nature. 2013. PMID: 23584588 Free PMC article. - Are bacteriophage defence and virulence two sides of the same coin in Campylobacter jejuni?
Louwen R, van Baarlen P. Louwen R, et al. Biochem Soc Trans. 2013 Dec;41(6):1475-81. doi: 10.1042/BST20130127. Biochem Soc Trans. 2013. PMID: 24256240 Review.
Cited by
- In silico Analysis Suggests Common Appearance of scaRNAs in Type II Systems and Their Association With Bacterial Virulence.
Guzina J, Chen WH, Stankovic T, Djordjevic M, Zdobnov E, Djordjevic M. Guzina J, et al. Front Genet. 2018 Oct 17;9:474. doi: 10.3389/fgene.2018.00474. eCollection 2018. Front Genet. 2018. PMID: 30386377 Free PMC article. - Characterization of CRISPR-Cas Systems in Serratia marcescens Isolated from Rhynchophorus ferrugineus (Olivier, 1790) (Coleoptera: Curculionidae).
Scrascia M, D'Addabbo P, Roberto R, Porcelli F, Oliva M, Calia C, Dionisi AM, Pazzani C. Scrascia M, et al. Microorganisms. 2019 Sep 19;7(9):368. doi: 10.3390/microorganisms7090368. Microorganisms. 2019. PMID: 31546915 Free PMC article. - Elevated temperature alters proteomic responses of individual organisms within a biofilm community.
Mosier AC, Li Z, Thomas BC, Hettich RL, Pan C, Banfield JF. Mosier AC, et al. ISME J. 2015 Jan;9(1):180-94. doi: 10.1038/ismej.2014.113. Epub 2014 Jul 22. ISME J. 2015. PMID: 25050524 Free PMC article. - Widespread CRISPR repeat-like RNA regulatory elements in CRISPR-Cas systems.
Shmakov SA, Barth ZK, Makarova KS, Wolf YI, Brover V, Peters JE, Koonin EV. Shmakov SA, et al. bioRxiv [Preprint]. 2023 Mar 3:2023.03.03.530964. doi: 10.1101/2023.03.03.530964. bioRxiv. 2023. PMID: 37090614 Free PMC article. Updated. Preprint. - Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection.
Piattelli E, Peltier J, Soutourina O. Piattelli E, et al. Genes (Basel). 2020 Oct 16;11(10):1209. doi: 10.3390/genes11101209. Genes (Basel). 2020. PMID: 33081172 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- P51 OD011132/OD/NIH HHS/United States
- R01-AI110701/AI/NIAID NIH HHS/United States
- R01 AI110701/AI/NIAID NIH HHS/United States
- U54-AI057157/AI/NIAID NIH HHS/United States
- U54 AI057157/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources