Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation - PubMed (original) (raw)
. 2014 May 20;111(20):7319-24.
doi: 10.1073/pnas.1324151111. Epub 2014 Apr 28.
Teng Fei 2, Yiwen Chen 3, Tiantian Li 1, Yanfei Gao 4, Xiaodong Wang 1, Tong Sun 1, Christopher J Sweeney 1, Gwo-Shu Mary Lee 1, Shaoyong Chen 4, Steven P Balk 4, Xiaole Shirley Liu 3, Myles Brown 2, Philip W Kantoff 5
Affiliations
- PMID: 24778216
- PMCID: PMC4034202
- DOI: 10.1073/pnas.1324151111
Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation
Chen-Lin Hsieh et al. Proc Natl Acad Sci U S A. 2014.
Abstract
The androgen receptor (AR) is a key factor that regulates the behavior and fate of prostate cancer cells. The AR-regulated network is activated when AR binds enhancer elements and modulates specific enhancer-promoter looping. Kallikrein-related peptidase 3 (KLK3), which codes for prostate-specific antigen (PSA), is a well-known AR-regulated gene and its upstream enhancers produce bidirectional enhancer RNAs (eRNAs), termed KLK3e. Here, we demonstrate that KLK3e facilitates the spatial interaction of the KLK3 enhancer and the KLK2 promoter and enhances long-distance KLK2 transcriptional activation. KLK3e carries the core enhancer element derived from the androgen response element III (ARE III), which is required for the interaction of AR and Mediator 1 (Med1). Furthermore, we show that KLK3e processes RNA-dependent enhancer activity depending on the integrity of core enhancer elements. The transcription of KLK3e was detectable and its expression is significantly correlated with KLK3 (R(2) = 0.6213, P < 5 × 10(-11)) and KLK2 (R(2) = 0.5893, P < 5 × 10(-10)) in human prostate tissues. Interestingly, RNAi silencing of KLK3e resulted in a modest negative effect on prostate cancer cell proliferation. Accordingly, we report that an androgen-induced eRNA scaffolds the AR-associated protein complex that modulates chromosomal architecture and selectively enhances AR-dependent gene expression.
Keywords: KLK3e/AR/Med1 complex; chromosomal looping.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
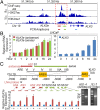
The KLK3 enhancer produces lncRNAs. (A) The chromatin status in the vicinity of KLK3. ChIP-seq datasets of AR, H3K27ac, and H3K4me1 (19). (B) RT-qPCR analyses of expression of antisense KLK3 eRNA (red bar), sense KLK3 eRNA (green bar), and mature KLK3 (blue bar) upon DHT (10 nM) treatment in a time-dependent manner in LNCaP cells. (C, Upper) A diagram of functional ARE positions that are relative to the KLK3 transcription starting site (TSS) adapted from ref. . (Lower) RT-qPCR analyses of the expression of segments of KLK3e with DHT (10 nM) treatment in LNCaP cells. The regions of LNA probes against the antisense and sense KLK3e were as indicated. Primer sets for qPCR were designed to specifically target either antisense (primers AS1–AS4) or sense strand (primer S1–S7) of KLK3e as shown. The right electrophoresis images show the PCR products of antisense (2,230 bp) and sense (2,170 bp) KLK3e. All data analyzed by RT-qPCR were normalized to the expression level in vehicle (−). Data are shown as mean ± SD (n ≥ 3).
Fig. 2.
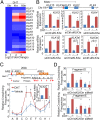
The regulatory function of KLK3e on the KLK locus. (A) A heat map of the KLK transcripts upon 4 and 16 h of DHT exposure in LNCaP cells (21). Genes are listed as their order in the kallikrein loci from centromere (Top) to telomere (Bottom). (B, Upper) Schematic drawing indicates the relative position of AR-regulated KLK genes and KLK3e on the chromosome 19. (Lower) LNCaP cells were treated with vehicle (−) or DHT (10 nM) and 50 nM of nonsilencing control (siCtrl) or KLK3e siRNA (siKLK3e); lowercase “e” denoted as eRNA. Efficacy and effects of KLK3e depletion on KLK genes were assessed by RT-qPCR. (C, Upper) Schematic graph shows the KLK3/2 gene loci. (Lower) LNCaP cells were treated with vehicle or DHT for 4 h. KLK2 mRNA expression was assessed by RT-qPCR (Inset). The relative cross-linking frequency between the anchor region (ARE III) and distal fragments (shaded bars) was determined by qPCR and normalized to the control region (fragment A). (D) The relative cross-linking frequency between the anchor region D and E region was determined by qPCR and normalized to the A region. All data analyzed by RT-qPCR or qPCR were normalized to the expression level in vehicle and are shown as mean ± SD (n ≥ 3), and *P < 0.05.
Fig. 3.
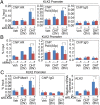
The KLK3e/AR/Med1 ribonucleoprotein complex transcriptionally regulates target promoters. Chromatin immunoprecipitation (ChIP) of AR or Med1 or activated Pol II (S5p) was performed in KLK3e- or Med1-depleted LNCaP cells with or without DHT (10 nM) for 2 or 8 h. (A–C) qPCR was conducted to measure the action of AR, Pol II (S5p), and Med1 at the KLK2 promoter. Data are shown as mean ± SD (n ≥ 3) and *P < 0.05.
Fig. 4.
The core enhancer element renders KLK3e RNA-dependent enhancer activity. (A, Upper) Schematic representation shows the insertion in KLK2 promoter driven luciferase reporter. (Lower) Reporter assay analyses were performed with or without insertion of full-length (S1–S7), flipped (S7–S1), and ARE deleted (S2–S7) of sense KLK3e. (B) Reporter analysis of cotransfecting the reporter with independent overexpressing vector as indicated. (C) The effects of siKLK3e (Left) or siMed1 (Right) on the KLK2 promoter-driven Luciferase activity. Luciferase activity was measured in the presence of DHT (10 nM) for 24 h. All data shown are mean ± SD of at least three independent experiments. *P < 0.05; firefly luciferase/Renilla luciferase (FL/RL).
Fig. 5.
KLK3e selectively enhances AR-regulated gene expression. (A) LNCaP cells were treated with vehicle or DHT (10 nM) and/or siCtrl or siKLK3e. The effect of siKLK3e on AR-regulated gene expression was determined by RT-qPCR. (B) Correlation between KLK3e (S1) and target gene expression in vivo. All data were normalized for GAPDH by the differences in threshold cycles (∆_C_T) from the normal human prostate gland (n = 4), primary (n = 11) and metastatic (n = 32) prostate tumors (25). (C) LNCaP cells were cultured in 5% charcoal/dextran-treated FBS (C-FBS) medium, followed by siRNA knockdown of scrambled control (siCtrl) or KLK3e (siKLK3e) with ethanol (veh) or DHT (10 nM) treatment. Cell growth was measured by WST1 assay. Data represent mean ± SD (n ≥ 3), and *P < 0.05. (D) A proposed model for functional activities of the eRNA/AR/Med1 complex in DHT-induced cis (solid line) and potential trans (dish line) gene activation events.
Similar articles
- Long terminal repeats act as androgen-responsive enhancers for the PSA-kallikrein locus.
Lawrence MG, Stephens CR, Need EF, Lai J, Buchanan G, Clements JA. Lawrence MG, et al. Endocrinology. 2012 Jul;153(7):3199-210. doi: 10.1210/en.2012-1267. Epub 2012 May 17. Endocrinology. 2012. PMID: 22597536 - Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells.
Andreu-Vieyra C, Lai J, Berman BP, Frenkel B, Jia L, Jones PA, Coetzee GA. Andreu-Vieyra C, et al. Mol Cell Biol. 2011 Dec;31(23):4648-62. doi: 10.1128/MCB.05934-11. Epub 2011 Oct 3. Mol Cell Biol. 2011. PMID: 21969603 Free PMC article. - Androgen receptor enhancer usage and the chromatin regulatory landscape in human prostate cancers.
Stelloo S, Bergman AM, Zwart W. Stelloo S, et al. Endocr Relat Cancer. 2019 May;26(5):R267-R285. doi: 10.1530/ERC-19-0032. Endocr Relat Cancer. 2019. PMID: 30865928 Review. - Androgen Receptor-Mediated Transcription in Prostate Cancer.
Özturan D, Morova T, Lack NA. Özturan D, et al. Cells. 2022 Mar 5;11(5):898. doi: 10.3390/cells11050898. Cells. 2022. PMID: 35269520 Free PMC article. Review.
Cited by
- Epigenetics of neural differentiation: Spotlight on enhancers.
Giacoman-Lozano M, Meléndez-Ramírez C, Martinez-Ledesma E, Cuevas-Diaz Duran R, Velasco I. Giacoman-Lozano M, et al. Front Cell Dev Biol. 2022 Oct 13;10:1001701. doi: 10.3389/fcell.2022.1001701. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313573 Free PMC article. Review. - Functional mapping of androgen receptor enhancer activity.
Huang CF, Lingadahalli S, Morova T, Ozturan D, Hu E, Yu IPL, Linder S, Hoogstraat M, Stelloo S, Sar F, van der Poel H, Altintas UB, Saffarzadeh M, Le Bihan S, McConeghy B, Gokbayrak B, Feng FY, Gleave ME, Bergman AM, Collins C, Hach F, Zwart W, Emberly E, Lack NA. Huang CF, et al. Genome Biol. 2021 May 11;22(1):149. doi: 10.1186/s13059-021-02339-6. Genome Biol. 2021. PMID: 33975627 Free PMC article. - CRISPR Screening of Transcribed Super-Enhancers Identifies Drivers of Triple-Negative Breast Cancer Progression.
Lewis MW, King CM, Wisniewska K, Regner MJ, Coffey A, Kelly MR, Mendez-Giraldez R, Davis ES, Phanstiel DH, Franco HL. Lewis MW, et al. Cancer Res. 2024 Nov 4;84(21):3684-3700. doi: 10.1158/0008-5472.CAN-23-3995. Cancer Res. 2024. PMID: 39186674 - Enhancer RNA (eRNA) in Human Diseases.
Wang Y, Zhang C, Wang Y, Liu X, Zhang Z. Wang Y, et al. Int J Mol Sci. 2022 Sep 30;23(19):11582. doi: 10.3390/ijms231911582. Int J Mol Sci. 2022. PMID: 36232885 Free PMC article. Review. - Super-Enhancers at the Nanog Locus Differentially Regulate Neighboring Pluripotency-Associated Genes.
Blinka S, Reimer MH Jr, Pulakanti K, Rao S. Blinka S, et al. Cell Rep. 2016 Sep 27;17(1):19-28. doi: 10.1016/j.celrep.2016.09.002. Cell Rep. 2016. PMID: 27681417 Free PMC article.
References
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25(2):276–308. - PubMed
- Nelson PS. Molecular states underlying androgen receptor activation: A framework for therapeutics targeting androgen signaling in prostate cancer. J Clin Oncol. 2012;30(6):644–646. - PubMed
- Hendriksen PJM, et al. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Res. 2006;66(10):5012–5020. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous