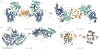Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity - PubMed (original) (raw)
Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity
James K Nuñez et al. Nat Struct Mol Biol. 2014 Jun.
Abstract
The initial stage of CRISPR-Cas immunity involves the integration of foreign DNA spacer segments into the host genomic CRISPR locus. The nucleases Cas1 and Cas2 are the only proteins conserved among all CRISPR-Cas systems, yet the molecular functions of these proteins during immunity are unknown. Here we show that Cas1 and Cas2 from Escherichia coli form a stable complex that is essential for spacer acquisition and determine the 2.3-Å-resolution crystal structure of the Cas1-Cas2 complex. Mutations that perturb Cas1-Cas2 complex formation disrupt CRISPR DNA recognition and spacer acquisition in vivo. Active site mutants of Cas2, unlike those of Cas1, can still acquire new spacers, thus indicating a nonenzymatic role of Cas2 during immunity. These results reveal the universal roles of Cas1 and Cas2 and suggest a mechanism by which Cas1-Cas2 complexes specify sites of CRISPR spacer integration.
Figures
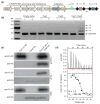
Figure 1. Cas1 and Cas2 associate to form a complex
(a) Representation of the CRISPR–Cas locus of E. coli K12. The 33-bp spacers (squares) are separated by 28-bp repeats (black diamonds). The half arrows flanking the leader and repeat-spacer arrays represent the positions of the primers used for PCR amplification in the spacer acquisition assays in BL21-AI cells. (b) Agarose gel of the PCR amplified CRISPR-I locus of BL21-AI cells after induced expression of empty vector, Cas1, Cas2 or Cas1+Cas2. Distinct bands represent the number of repeat-spacer arrays additions into the genomic parental CRISPR locus. (c) FLAG- and HA-immunoprecipitations in lysates overexpressing Cas1 only, Cas2 only or both. (d) ITC trace of Cas1 injection into a Cas2-containing cell. The reported N and _K_d values are averages of three independent experiments.
Figure 2. Crystal structure of the Cas1–Cas2 complex
(a) The overall structure consists of a Cas2 dimer (yellow and orange) and two Cas1 dimers (a-d, blue and teal). (b) Superposition of the Cas1a-b dimer with the previously determined E. coli Cas1 structure (gray, PDB 3NKD). The dashed orange circle highlights the conformational change observed in the a-helical domain of Cas1a. (c) Superposition of the Cas2 dimer in the complex with the previously determined E. coli Cas2 structure (gray, PDB 4MAK). The arrows point to the last resolved residue in the 4MAK structure. The N and C indicate the termini of each monomer; root-mean-square deviations (r.m.s.d.) of the superpositions are indicated.
Figure 3. Disruption of complex formation affects spacer acquisition in vivo
(a) A close-up view of the Cas1a–Cas2 protein-protein interface, with annotations for the residues involved in electrostatic interactions. (b) View of the ordered C-terminal tail of Cas1a with the electron density mesh contoured at 1.0 sigma. (c–e) Agarose gels of in vivo acquisition assays with mutations of Cas1 and Cas2 at the C-termini (c) and the electrostatic interface (d,e). (f) Western blot of FLAG immunoprecipitations in BL21-AI cells expressing Cas1-FLAG and Cas2-HA or various mutations of Cas1 and Cas2. Despite the low expression of Cas2 E65R, we still detect its co-elution with Cas1.
Figure 4. Complex formation is required for CRISPR DNA recognition
(a,b) Close-up views of the Cas1 and Cas2 active sites with stick representations for the conserved residues mutated in vivo. In the middle is a general view of the active sites in the complex, highlighted in red. (c,d) Acquisition assays of active site residue mutations of Cas1 and Cas2. (e) Western blot of fractions in the biotinylated DNA affinity precipitations. The cartoon representations are the DNA constructs used with the 5′-biotin labels (stars). W.C.L. refers to the whole cell lysate. (f) Western blot of the DNA affinity precipitations in BL21-AI lysates from overexpression of Cas1-FLAG only, Cas2-HA only or both. (g) FLAG IP in lysates from overexpression of Cas1-FLAG and Cas2 E9A-HA. (h) DNA affinity precipitations in the same lysates as in (g) using the same DNA constructs as in (e).
Similar articles
- Harnessing CRISPR-Cas adaptation for RNA recording and beyond.
Oh GS, An S, Kim S. Oh GS, et al. BMB Rep. 2024 Jan;57(1):40-49. doi: 10.5483/BMBRep.2023-0050. BMB Rep. 2024. PMID: 38053290 Free PMC article. Review. - How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Xiao Y, Ng S, Nam KH, Ke A. Xiao Y, et al. Nature. 2017 Oct 5;550(7674):137-141. doi: 10.1038/nature24020. Epub 2017 Sep 4. Nature. 2017. PMID: 28869593 Free PMC article. - Structural plasticity and in vivo activity of Cas1 from the type I-F CRISPR-Cas system.
Wilkinson ME, Nakatani Y, Staals RH, Kieper SN, Opel-Reading HK, McKenzie RE, Fineran PC, Krause KL. Wilkinson ME, et al. Biochem J. 2016 Apr 15;473(8):1063-72. doi: 10.1042/BCJ20160078. Epub 2016 Feb 29. Biochem J. 2016. PMID: 26929403 - CRISPR Immunological Memory Requires a Host Factor for Specificity.
Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA. Nuñez JK, et al. Mol Cell. 2016 Jun 16;62(6):824-833. doi: 10.1016/j.molcel.2016.04.027. Epub 2016 May 19. Mol Cell. 2016. PMID: 27211867 - CRISPR-Cas adaptation in Escherichia coli.
Mitić D, Bolt EL, Ivančić-Baće I. Mitić D, et al. Biosci Rep. 2023 Mar 31;43(3):BSR20221198. doi: 10.1042/BSR20221198. Biosci Rep. 2023. PMID: 36809461 Free PMC article. Review.
Cited by
- Harnessing CRISPR-Cas adaptation for RNA recording and beyond.
Oh GS, An S, Kim S. Oh GS, et al. BMB Rep. 2024 Jan;57(1):40-49. doi: 10.5483/BMBRep.2023-0050. BMB Rep. 2024. PMID: 38053290 Free PMC article. Review. - DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins.
Lisitskaya L, Aravin AA, Kulbachinskiy A. Lisitskaya L, et al. Nat Commun. 2018 Dec 4;9(1):5165. doi: 10.1038/s41467-018-07449-7. Nat Commun. 2018. PMID: 30514832 Free PMC article. Review. - Editor's cut: DNA cleavage by CRISPR RNA-guided nucleases Cas9 and Cas12a.
Swartjes T, Staals RHJ, van der Oost J. Swartjes T, et al. Biochem Soc Trans. 2020 Feb 28;48(1):207-219. doi: 10.1042/BST20190563. Biochem Soc Trans. 2020. PMID: 31872209 Free PMC article. Review. - CRISPR-Cas systems are present predominantly on chromosome and its relationship with MEGs in Vibrio species.
Zhang E, Zhou W, Zhou J, He Z, Zhou Y, Han J, Qu D. Zhang E, et al. Arch Microbiol. 2021 Dec 25;204(1):76. doi: 10.1007/s00203-021-02656-1. Arch Microbiol. 2021. PMID: 34953139 - Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci.
Shiimori M, Garrett SC, Graveley BR, Terns MP. Shiimori M, et al. Mol Cell. 2018 Jun 7;70(5):814-824.e6. doi: 10.1016/j.molcel.2018.05.002. Epub 2018 Jun 7. Mol Cell. 2018. PMID: 29883605 Free PMC article.
References
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–66. - PubMed
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. - PubMed
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–61. - PubMed
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials