Histone variant H3.3 is an essential maternal factor for oocyte reprogramming - PubMed (original) (raw)
Histone variant H3.3 is an essential maternal factor for oocyte reprogramming
Duancheng Wen et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Mature oocyte cytoplasm can reprogram somatic cell nuclei to the pluripotent state through a series of sequential events including protein exchange between the donor nucleus and ooplasm, chromatin remodeling, and pluripotency gene reactivation. Maternal factors that are responsible for this reprogramming process remain largely unidentified. Here, we demonstrate that knockdown of histone variant H3.3 in mouse oocytes results in compromised reprogramming and down-regulation of key pluripotency genes; and this compromised reprogramming for developmental potentials and transcription of pluripotency genes can be rescued by injecting exogenous H3.3 mRNA, but not H3.2 mRNA, into oocytes in somatic cell nuclear transfer embryos. We show that maternal H3.3, and not H3.3 in the donor nucleus, is essential for successful reprogramming of somatic cell nucleus into the pluripotent state. Furthermore, H3.3 is involved in this reprogramming process by remodeling the donor nuclear chromatin through replacement of donor nucleus-derived H3 with de novo synthesized maternal H3.3 protein. Our study shows that H3.3 is a crucial maternal factor for oocyte reprogramming and provides a practical model to directly dissect the oocyte for its reprogramming capacity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
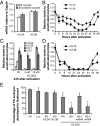
Fig. 1.
H3.3 knockdown compromises embryonic development of PA embryos. (A) H3.3 transcript levels in MII and enucleated oocytes. For all panels, error bars indicate average ± SD (n ≥ 3). (B) Dynamics of H3.3 transcript levels in PA embryos. Data are mean expression relative to Gapdh with unactivated oocytes normalized to 1. (C) Transcript analysis of H3.1, H3.2, and H3.3 levels after H3.3 siRNA (siH3.3) treatment and 24 h of activation. Data are mean expression relative to GAPDH with unactivated oocytes normalized to 1. H3.3KD embryos were subject to injection of 4 μM or 10 μM siH3.3 as indicated. (D) Dynamics of H3.3 transcript levels in oocytes treated with siH3.3 (four sets, 10 μM each) 1 h before activation. (E) Developmental potential of PA embryos. Control, unmanipulated PA embryos; KD-AA (after activation), siH3.3 (four sets, 4 µM each) 12 h after activation; KD-BA (before activation), siH3.3 (four sets, 4 µM each) before activation; KD+H3.2mRNA, H3.2mRNA (30 ng/µL) after siH3.3 (four sets, 4 µM each) injection; luciferase, siRNA against luciferase (16 µM); KD-H3.3A, siH3.3A only (two sets, 4 µM each); KD-H3.3B, siH3.3B only (two sets, 4 µM each); KD+H3.3mRNA, H3.3mRNA (30 ng/µL) after siH3.3 (four sets, 4 µM each) injection. Data were analyzed with χ_2_ test, and statistical significance was determined compared with luciferase (*P < 0.01). Error bars indicate average ± SD (n ≥ 3).
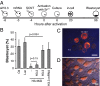
Fig. 2.
Compromised reprogramming in H3.3KD SCNT embryos. (A) Schematic illustration of siH3.3 injection into oocytes and SCNT. (B) Developmental potential of control and H3.3KD cumulus SCNT embryos exogenously expressing H3.2 or H3.3. Embryos were treated as described for PA embryos in Fig. 1, with all injections occurring before nuclear transfer. Data were analyzed with the χ2 test. Error bars indicate average ± SD (n ≥ 3). (C) SCNT blastocysts produced by transfer of H3.3KO embryonic fibroblast nuclei into WT enucleated oocytes. WT MEFs from the same chimeric embryos were also used as control. (D) ES cell lines (ntESCs) established from H3.3KO nuclear transfer blastocysts. EYFP and mCherry indicate the deletion of H3.3B gene. (Scale bar: 100 µm.)
Fig. 3.
Reactivation of pluripotency-associated genes is maternal H3.3-dependent in SCNT embryos. (A) Schematic illustration of blastomere injection and embryo culture. (B) Injection of Oct4-EGFP MEFs labeled with H2B-mCherry into one blastomere of a two-cell PA embryo and (C) culture to morula stage. (Scale bar: 100 µm.) (D and E) Injection of Oct4-EGFP MEFs into the blastomeres of H3.3KD two-cell embryos and control (D) or luciferase siRNA injected embryos (E). (F) Injection of Oct4-EGFP ES cells into the blastomeres of H3.3KD 2-cell embryos. (G) RNA-seq analysis of control and H3.3KD four-cell stage embryos exogenously expressing H3.2 or H3.3. (H) RNA-seq analysis of differential expression upon H3.3KD that was rescued by exogenous expression of H3.3. (I) RNA-seq analysis of 26 pluripotency genes (expressed in control SCNT embryos) in H3.3KD SCNT embryos. (J) Heat map of the 26 pluripotency genes in WT SCNT, luciferase-injected, H3.3KD, H3.3-addback, and H3.2-addback SCNT embryos at four-cell stage. (K) The average expression of the 26 pluripotency genes in control and H3.3KD embryos exogenously expressing H3.2 or H3.3.
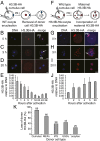
Fig. 4.
Maternal H3.3 replaces donor nucleus-derived H3 in the nuclei of SCNT embryos. (A_–_E) Embryos were constructed by injecting an H3.3B-HA cumulus nucleus into a WT B6D2F1 enucleated oocyte (A), allowing us to monitor the change of donor nucleus-derived H3.3 in SCNT embryo by using immunofluorescence (B_–_E). Donor H3.3B-HA, indicated by white arrow, was undetectable by the two-cell stage (20 h after activation). PB, polar body. (E) Dynamics of H3.3B-HA removal from donor nuclei. Data represent average HA intensities relative to DNA at various time points, with error representing SD (n ≥ 5). (F_–_J) Embryos were constructed by injecting a WT cumulus nucleus into an H3.3B-HA enucleated oocyte (F), allowing us to monitor the incorporation of maternal H3.3 into the donor nucleus using immunofluorescence (G_–_J). Maternal H3.3B-HA accumulated in the cytoplasm (H, white arrow) before incorporation into the donor nucleus at the two-cell stage (I). (J) Dynamics of maternal H3.3B-HA incorporation into SCNT embryos. Data represent average HA intensities relative to DNA at various time points, with error representing SD (n ≥ 5). (Scale bar: 20 µm.) (K) H3.3B-HA cumulus, MEF, iPS, ES cell, and oocyte nuclei were used for nuclear transfer and H3.3B-HA signal in the nuclei of the NT embryos was measured at 0 h and 20 h after activation. Data represent the percentage of reduced H3.3B-HA intensities in the nuclei of the embryos 20 h after activation relative to 0 h [(1 − HA20h/HA0h) × 100%].
Similar articles
- H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos.
Wen D, Banaszynski LA, Rosenwaks Z, Allis CD, Rafii S. Wen D, et al. Nucleus. 2014 Sep-Oct;5(5):369-75. doi: 10.4161/nucl.36231. Nucleus. 2014. PMID: 25482190 Review. - Dynamic replacement of H3.3 affects nuclear reprogramming in early bovine SCNT embryos.
Wang Y, Li Y, Luan D, Kang J, He R, Zhang Y, Quan F. Wang Y, et al. Theriogenology. 2020 Sep 15;154:43-52. doi: 10.1016/j.theriogenology.2020.05.031. Epub 2020 May 23. Theriogenology. 2020. PMID: 32480063 - Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming.
Jullien J, Miyamoto K, Pasque V, Allen GE, Bradshaw CR, Garrett NJ, Halley-Stott RP, Kimura H, Ohsumi K, Gurdon JB. Jullien J, et al. Mol Cell. 2014 Aug 21;55(4):524-36. doi: 10.1016/j.molcel.2014.06.024. Epub 2014 Jul 24. Mol Cell. 2014. PMID: 25066233 Free PMC article. - Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process?
Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Jullien J, et al. Nat Rev Mol Cell Biol. 2011 Jun 23;12(7):453-9. doi: 10.1038/nrm3140. Nat Rev Mol Cell Biol. 2011. PMID: 21697902 Free PMC article. Review.
Cited by
- Reprogramming of cell fate: epigenetic memory and the erasure of memories past.
Nashun B, Hill PW, Hajkova P. Nashun B, et al. EMBO J. 2015 May 12;34(10):1296-308. doi: 10.15252/embj.201490649. Epub 2015 Mar 27. EMBO J. 2015. PMID: 25820261 Free PMC article. Review. - The Histone Variant H3.3 Is Enriched at Drosophila Amplicon Origins but Does Not Mark Them for Activation.
Paranjape NP, Calvi BR. Paranjape NP, et al. G3 (Bethesda). 2016 Jun 1;6(6):1661-71. doi: 10.1534/g3.116.028068. G3 (Bethesda). 2016. PMID: 27172191 Free PMC article. - Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Harada A, Maehara K, Ono Y, Taguchi H, Yoshioka K, Kitajima Y, Xie Y, Sato Y, Iwasaki T, Nogami J, Okada S, Komatsu T, Semba Y, Takemoto T, Kimura H, Kurumizaka H, Ohkawa Y. Harada A, et al. Nat Commun. 2018 Apr 11;9(1):1400. doi: 10.1038/s41467-018-03845-1. Nat Commun. 2018. PMID: 29643389 Free PMC article. - Maternal histone variants and their chaperones promote paternal genome activation and boost somatic cell reprogramming.
Yang P, Wu W, Macfarlan TS. Yang P, et al. Bioessays. 2015 Jan;37(1):52-9. doi: 10.1002/bies.201400072. Epub 2014 Oct 18. Bioessays. 2015. PMID: 25328107 Free PMC article. Review. - CHD1 Regulates Deposition of Histone Variant H3.3 During Bovine Early Embryonic Development.
Zhang K, Rajput SK, Wang S, Folger JK, Knott JG, Smith GW. Zhang K, et al. Biol Reprod. 2016 Jun;94(6):140. doi: 10.1095/biolreprod.116.138693. Epub 2016 May 11. Biol Reprod. 2016. PMID: 27170440 Free PMC article.
References
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. - PubMed
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. - PubMed
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394(6691):369–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2 HL101846/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01HL097797/HL/NHLBI NIH HHS/United States
- U54CA163167/CA/NCI NIH HHS/United States
- R01 HL097797/HL/NHLBI NIH HHS/United States
- R01 HL119872/HL/NHLBI NIH HHS/United States
- RC2HL101846/HL/NHLBI NIH HHS/United States
- U54 CA163167/CA/NCI NIH HHS/United States
- R01 DK095039/DK/NIDDK NIH HHS/United States
- R01HL119872/HL/NHLBI NIH HHS/United States
- R01DK095039/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases