Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe - PubMed (original) (raw)
. 2014 May 8;10(5):e1004353.
doi: 10.1371/journal.pgen.1004353. eCollection 2014 May.
Meredith L Carpenter 1, Andres Moreno-Estrada 1, Brenna M Henn 1, Peter A Underhill 1, Federico Sánchez-Quinto 2, Ilenia Zara 3, Maristella Pitzalis 4, Carlo Sidore 5, Fabio Busonero 6, Andrea Maschio 6, Andrea Angius 6, Chris Jones 3, Javier Mendoza-Revilla 7, Georgi Nekhrizov 8, Diana Dimitrova 8, Nikola Theodossiev 9, Timothy T Harkins 10, Andreas Keller 11, Frank Maixner 12, Albert Zink 12, Goncalo Abecasis 13, Serena Sanna 4, Francesco Cucca 4, Carlos D Bustamante 1
Affiliations
- PMID: 24809476
- PMCID: PMC4014435
- DOI: 10.1371/journal.pgen.1004353
Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe
Martin Sikora et al. PLoS Genet. 2014.
Abstract
Genome sequencing of the 5,300-year-old mummy of the Tyrolean Iceman, found in 1991 on a glacier near the border of Italy and Austria, has yielded new insights into his origin and relationship to modern European populations. A key finding of that study was an apparent recent common ancestry with individuals from Sardinia, based largely on the Y chromosome haplogroup and common autosomal SNP variation. Here, we compiled and analyzed genomic datasets from both modern and ancient Europeans, including genome sequence data from over 400 Sardinians and two ancient Thracians from Bulgaria, to investigate this result in greater detail and determine its implications for the genetic structure of Neolithic Europe. Using whole-genome sequencing data, we confirm that the Iceman is, indeed, most closely related to Sardinians. Furthermore, we show that this relationship extends to other individuals from cultural contexts associated with the spread of agriculture during the Neolithic transition, in contrast to individuals from a hunter-gatherer context. We hypothesize that this genetic affinity of ancient samples from different parts of Europe with Sardinians represents a common genetic component that was geographically widespread across Europe during the Neolithic, likely related to migrations and population expansions associated with the spread of agriculture.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
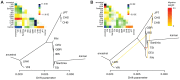
Figure 1. Geographic origin of ancient samples and ADMIXTURE results.
(A) Map of Europe indicating the discovery sites for each of the ancient samples used in this study. (B) Ancestral population clusters inferred using ADMIXTURE on the HGDP dataset, for k = 6 ancestral clusters. The width of the bars of the ancient samples was increased to aid visualization.
Figure 2. Allele sharing and D-tests with whole-genome datasets.
(A) Normalized derived allele sharing rate of the Iceman with Eurasian whole genomes from Complete Genomics. Each circle represents the rate of sharing with a particular genome, grouped by population of origin. Positions on the y-axis have added jitter for ease of visualization. Populations with EUR suffix correspond to the European ancestry tracts of individuals of populations with known European admixture (ASW, MXL). Due to differences in admixture proportions among individuals from those populations, the total number of observations varies between individuals, indicated by the size of the circles. (B) D-test results for the ancient samples compared to populations from the 1000 Genomes project and Sardinia. Each panel shows results for a particular ancient sample, grouped by cultural context. Diamonds indicates the value of the D-statistic for a single D-test involving the ancient sample and a pair of modern populations, shown on the left and right of the panels. Significance at Z = 3 is indicated with filled diamonds, and the line shows the corresponding standard error of the D-statistic. Plot colors indicate different pairs of geographic regions within Europe (blue: Europe S/Europe N; green: Europe S/Europe S).
Figure 3. Results of TreeMix analysis of the Iceman with 1000G/Sardinia.
Shown are maximum-likelihood trees and the matrices of pairwise residuals (inset) for a model allowing (A) m = 0 and (B) m = 3 mixture events. Large positive values in the residual matrix indicate a poor fit for the respective pair of populations. Edges representing mixture events are colored according to weight of the inferred edge.
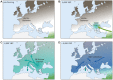
Figure 4. A simplified model for recent demographic history of Europeans.
The panels indicate a possible demographic scenario consistent with the observed signals. (A) Mesolithic HGs present in mainland Europe prior to the arrival of agriculture. (B) Initial spread of farming from the Middle East beginning from 7,000 YBP into SE Europe. (C) Expansion of farming to N Europe from SE European gene pool and establishment of main S-N gradient of genetic diversity. Wave of migration also reaches Sardinia. (D) Continuous population expansion and admixture with local HGs as well as additional migrations continues to shape genetic diversity in mainland Europe, but Sardinia remains mostly isolated (IBD: isolation by distance).
Similar articles
- Whole mitochondrial DNA sequencing in Alpine populations and the genetic history of the Neolithic Tyrolean Iceman.
Coia V, Cipollini G, Anagnostou P, Maixner F, Battaggia C, Brisighelli F, Gómez-Carballa A, Destro Bisol G, Salas A, Zink A. Coia V, et al. Sci Rep. 2016 Jan 14;6:18932. doi: 10.1038/srep18932. Sci Rep. 2016. PMID: 26764605 Free PMC article. - High-coverage genome of the Tyrolean Iceman reveals unusually high Anatolian farmer ancestry.
Wang K, Prüfer K, Krause-Kyora B, Childebayeva A, Schuenemann VJ, Coia V, Maixner F, Zink A, Schiffels S, Krause J. Wang K, et al. Cell Genom. 2023 Aug 16;3(9):100377. doi: 10.1016/j.xgen.2023.100377. eCollection 2023 Sep 13. Cell Genom. 2023. PMID: 37719142 Free PMC article. - Complete mitochondrial genome sequence of the Tyrolean Iceman.
Ermini L, Olivieri C, Rizzi E, Corti G, Bonnal R, Soares P, Luciani S, Marota I, De Bellis G, Richards MB, Rollo F. Ermini L, et al. Curr Biol. 2008 Nov 11;18(21):1687-93. doi: 10.1016/j.cub.2008.09.028. Epub 2008 Oct 30. Curr Biol. 2008. PMID: 18976917 - The Current Situation of the Tyrolean Iceman.
Zink AR, Maixner F. Zink AR, et al. Gerontology. 2019;65(6):699-706. doi: 10.1159/000501878. Epub 2019 Sep 10. Gerontology. 2019. PMID: 31505504 Review. - Through 40,000 years of human presence in Southern Europe: the Italian case study.
Aneli S, Caldon M, Saupe T, Montinaro F, Pagani L. Aneli S, et al. Hum Genet. 2021 Oct;140(10):1417-1431. doi: 10.1007/s00439-021-02328-6. Epub 2021 Aug 19. Hum Genet. 2021. PMID: 34410492 Free PMC article. Review.
Cited by
- Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean.
Schroeder H, Ávila-Arcos MC, Malaspinas AS, Poznik GD, Sandoval-Velasco M, Carpenter ML, Moreno-Mayar JV, Sikora M, Johnson PL, Allentoft ME, Samaniego JA, Haviser JB, Dee MW, Stafford TW Jr, Salas A, Orlando L, Willerslev E, Bustamante CD, Gilbert MT. Schroeder H, et al. Proc Natl Acad Sci U S A. 2015 Mar 24;112(12):3669-73. doi: 10.1073/pnas.1421784112. Epub 2015 Mar 9. Proc Natl Acad Sci U S A. 2015. PMID: 25755263 Free PMC article. - Efficient analysis of large datasets and sex bias with ADMIXTURE.
Shringarpure SS, Bustamante CD, Lange K, Alexander DH. Shringarpure SS, et al. BMC Bioinformatics. 2016 May 23;17:218. doi: 10.1186/s12859-016-1082-x. BMC Bioinformatics. 2016. PMID: 27216439 Free PMC article. - The genetic prehistory of the Andean highlands 7000 years BP though European contact.
Lindo J, Haas R, Hofman C, Apata M, Moraga M, Verdugo RA, Watson JT, Viviano Llave C, Witonsky D, Beall C, Warinner C, Novembre J, Aldenderfer M, Di Rienzo A. Lindo J, et al. Sci Adv. 2018 Nov 8;4(11):eaau4921. doi: 10.1126/sciadv.aau4921. eCollection 2018 Nov. Sci Adv. 2018. PMID: 30417096 Free PMC article. - CT Scan of Thirteen Natural Mummies Dating Back to the XVI-XVIII Centuries: An Emerging Tool to Investigate Living Conditions and Diseases in History.
Petrella E, Piciucchi S, Feletti F, Barone D, Piraccini A, Minghetti C, Gruppioni G, Poletti V, Bertocco M, Traversari M. Petrella E, et al. PLoS One. 2016 Jun 29;11(6):e0154349. doi: 10.1371/journal.pone.0154349. eCollection 2016. PLoS One. 2016. PMID: 27355351 Free PMC article. - The Italian genome reflects the history of Europe and the Mediterranean basin.
Fiorito G, Di Gaetano C, Guarrera S, Rosa F, Feldman MW, Piazza A, Matullo G. Fiorito G, et al. Eur J Hum Genet. 2016 Jul;24(7):1056-62. doi: 10.1038/ejhg.2015.233. Epub 2015 Nov 11. Eur J Hum Genet. 2016. PMID: 26554880 Free PMC article.
References
- Seidler H, Bernhard W, Teschler-Nicola M, Platzer W, zur Nedden D, et al. (1992) Some anthropological aspects of the prehistoric Tyrolean ice man. Science 258: 455–457. - PubMed
- Murphy WA, zur Nedden D, Gostner P, Knapp R, Recheis W, et al. (2003) The Iceman: Discovery and Imaging. Radiology 226: 614–629. - PubMed
- Kutschera W, Golser R, Priller A, Rom W, Steier P, et al... (2000) Radiocarbon dating of equipment from the Iceman. In: Bortenschlager U-PMDS, Oeggl U-PDK, editors. The Iceman and his Natural Environment: Palaeobotanical Results (The Man in the Ice). Vienna: Springer. pp. 1–9.
- Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, et al. (2012) New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nature Communications 3: 698. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources