Phylogenomic reconstruction of archaeal fatty acid metabolism - PubMed (original) (raw)
Phylogenomic reconstruction of archaeal fatty acid metabolism
Daria V Dibrova et al. Environ Microbiol. 2014 Apr.
Abstract
While certain archaea appear to synthesize and/or metabolize fatty acids, the respective pathways still remain obscure. By analysing the genomic distribution of the key lipid-related enzymes, we were able to identify the likely components of the archaeal pathway of fatty acid metabolism, namely, a combination of the enzymes of bacterial-type β-oxidation of fatty acids [acyl-coenzyme A (CoA) dehydrogenase, enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase] with paralogs of the archaeal acetyl-CoA C-acetyltransferase, an enzyme of the mevalonate biosynthesis pathway. These three β-oxidation enzymes working in the reverse direction could potentially catalyse biosynthesis of fatty acids, with paralogs of acetyl-CoA C-acetyltransferase performing addition of C2 fragments. The presence in archaea of the genes for energy-transducing membrane enzyme complexes, such as cytochrome bc complex, cytochrome c oxidase and diverse rhodopsins, was found to correlate with the presence of the proposed system of fatty acid biosynthesis. We speculate that because these membrane complexes functionally depend on fatty acid chains, their genes could have been acquired via lateral gene transfer from bacteria only by those archaea that already possessed a system of fatty acid biosynthesis. The proposed pathway of archaeal fatty acid metabolism operates in extreme conditions and therefore might be of interest in the context of biofuel production and other industrial applications.
Figures
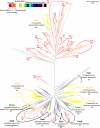
Figure 1. Schematic view of the phylogenetic tree for β-ketothiolases (acetoacetyl-CoA acyltransferases, COG0183)
Separate clades are labelled with the names of the experimentally characterized genes from model organisms. The names of E. coli genes are in black, names of B. subtilis genes are in green, names of human genes are in yellow. The branches of tree are colored according to the taxonomical identity of the source organisms; the color code is shown in the upper left corner. Clades with archaeal sequences are indicated by red arks or red arrows and named from A1 to AM_4 and from B1 to B4. The numbers underneath the clade labels indicate the reliability of respective branches (as tested using SH-like statistics implemented in PhyML). The bar in the top right corner represents the scale for the branch lengths (the expected number of substitutions per site). The tree has been constructed based on conserved blocks (total 227 positions) from 511 sequences. The sequences were aligned with Muscle (Edgar, 2004) and the alignment was manually edited using GeneDoc (Nicholas et al., 1997). The phylogenetic tree was constructed by PhyML (Guindon and Gascuel, 2003) with the SPR algorithm of tree construction. The tree was visualized with MEGA5 program (Tamura et al., 2011). The complete version of this tree is available in the Supplementary Materials as Figure S2.
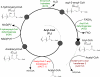
Figure 2. Proposed scheme of archaeal fatty acid biosynthesis
The synthesis of fatty acids in archaea is proposed to proceed through essentially the same steps as in FAS I or FAS II mechanisms (see Figure S3), but by using CoA instead of the acyl-carrier protein, as in the β-oxidation of fatty acids (see Figure S1). Enzymes that catalyze the reduction (HDH and ACD) and dehydratation (ECH) steps are proposed to be the same as those involved in the bacterial β-oxidation pathway, whereas initiation of the cycle and fatty acid chain elongation are proposed to be catalyzed by various paralogs of the archaeal acetyl-CoA C-acetyltransferase. As discussed in the text, the presence of several paralogs of the latter enzyme in many archaeal genomes (denoted here as AACT1 and AACT2) might reflect their specificity towards chains of particular lengths. As an example, enzymes of Aeropyrum pernix K1, proposed to catalyze the respective steps, are indicated by their genome locus tags (GenBank accession no. BA000002).
Similar articles
- Comparison of metabolic fluxes of cis-5-enoyl-CoA and saturated acyl-CoA through the beta-oxidation pathway.
Tserng KY, Chen LS, Jin SJ. Tserng KY, et al. Biochem J. 1995 Apr 1;307 ( Pt 1)(Pt 1):23-8. doi: 10.1042/bj3070023. Biochem J. 1995. PMID: 7717980 Free PMC article. - Peroxisomal beta-oxidation: enzymology and molecular biology.
Hashimoto T. Hashimoto T. Ann N Y Acad Sci. 1996 Dec 27;804:86-98. doi: 10.1111/j.1749-6632.1996.tb18610.x. Ann N Y Acad Sci. 1996. PMID: 8993538 Review. No abstract available. - Functions and organization of peroxisomal beta-oxidation.
Mannaerts GP, van Veldhoven PP. Mannaerts GP, et al. Ann N Y Acad Sci. 1996 Dec 27;804:99-115. doi: 10.1111/j.1749-6632.1996.tb18611.x. Ann N Y Acad Sci. 1996. PMID: 8993539 Review. No abstract available.
Cited by
- Life in hot acid: a genome-based reassessment of the archaeal order Sulfolobales.
Counts JA, Willard DJ, Kelly RM. Counts JA, et al. Environ Microbiol. 2021 Jul;23(7):3568-3584. doi: 10.1111/1462-2920.15189. Epub 2020 Sep 7. Environ Microbiol. 2021. PMID: 32776389 Free PMC article. - Uncovering major genomic features of essential genes in Bacteria and a methanogenic Archaea.
Grazziotin AL, Vidal NM, Venancio TM. Grazziotin AL, et al. FEBS J. 2015 Sep;282(17):3395-3411. doi: 10.1111/febs.13350. Epub 2015 Jul 14. FEBS J. 2015. PMID: 26084810 Free PMC article. - Genomic and transcriptomic dissection of Theionarchaea in marine ecosystem.
Cai M, Duan C, Zhang X, Pan J, Liu Y, Zhang C, Li M. Cai M, et al. Sci China Life Sci. 2022 Jun;65(6):1222-1234. doi: 10.1007/s11427-021-1996-x. Epub 2021 Oct 14. Sci China Life Sci. 2022. PMID: 34668130 - Enoyl-CoA hydratase mediates polyhydroxyalkanoate mobilization in Haloferax mediterranei.
Liu G, Cai S, Hou J, Zhao D, Han J, Zhou J, Xiang H. Liu G, et al. Sci Rep. 2016 Apr 7;6:24015. doi: 10.1038/srep24015. Sci Rep. 2016. PMID: 27052994 Free PMC article. - On the Archaeal Origins of Eukaryotes and the Challenges of Inferring Phenotype from Genotype.
Dey G, Thattai M, Baum B. Dey G, et al. Trends Cell Biol. 2016 Jul;26(7):476-485. doi: 10.1016/j.tcb.2016.03.009. Epub 2016 Jun 16. Trends Cell Biol. 2016. PMID: 27319280 Free PMC article. Review.
References
- Boucher Y, Kamekura M, Doolittle WF. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol Microbiol. 2004;52:515–527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous