Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B - PubMed (original) (raw)
Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B
Alexander J F Egan et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Bacteria surround their cytoplasmic membrane with an essential, stress-bearing peptidoglycan (PG) layer. Growing and dividing cells expand their PG layer by using membrane-anchored PG synthases, which are guided by dynamic cytoskeletal elements. In Escherichia coli, growth of the mainly single-layered PG is also regulated by outer membrane-anchored lipoproteins. The lipoprotein LpoB is required for the activation of penicillin-binding protein (PBP) 1B, which is a major, bifunctional PG synthase with glycan chain polymerizing (glycosyltransferase) and peptide cross-linking (transpeptidase) activities. Here, we report the structure of LpoB, determined by NMR spectroscopy, showing an N-terminal, 54-aa-long flexible stretch followed by a globular domain with similarity to the N-terminal domain of the prevalent periplasmic protein TolB. We have identified the interaction interface between the globular domain of LpoB and the noncatalytic UvrB domain 2 homolog domain of PBP1B and modeled the complex. Amino acid exchanges within this interface weaken the PBP1B-LpoB interaction, decrease the PBP1B stimulation in vitro, and impair its function in vivo. On the contrary, the N-terminal flexible stretch of LpoB is required to stimulate PBP1B in vivo, but is dispensable in vitro. This supports a model in which LpoB spans the periplasm to interact with PBP1B and stimulate PG synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
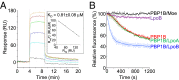
LpoB interacts with PBP1B and enhances the rate of its GTase activity. (A) LpoB–PBP1B interaction dynamics measured by SPR. LpoB was injected at concentrations of 0 (black line), 0.5 (light blue), 1 (dark blue), 2 (green), 3 (red), and 4 (orange) μM to a chip surface with immobilized PBP1B (
SI Appendix, Fig. S1_A_
shows control). The response at equilibrium _R_eq, normalized to the injected concentration C (_R_eq/C), plotted against _R_eq, yields a straight line with a slope of −_K_d−1 (Inset). The average _K_d value ± SD was determined from three independent experiments. (B) Continuous GTase assay for PBP1B, measuring consumption of fluorescently labeled substrate (dansyl-lipid II). PBP1B activity (red) is stimulated by LpoB (blue) but not by LpoA (green). The antibiotic moenomycin (Moe) inhibits PBP1B GTase activity (black). LpoB alone (purple) shows no activity. Each measurement is shown as mean ± SD of three independent experiments.
Fig. 2.
LpoB structure shows a globular domain with a flexible N-terminal region. (A) Ensemble of the 20 lowest-energy structures of LpoB(sol) (from Thr68 for clarity) determined by NMR spectroscopy (colors from cyan to red are from N to C terminus). The position of α-helices (H1–H4) and β-strands (β1–β4) are indicated on the structure and on the sequence below (helices and arrows). (B) Heteronuclear {1H}-15N-NOE values calculated from the intensity ratio between the saturated and the reference experiments. Errors on the intensity ratio were calculated from the signal-to-noise ratio of the NMR signal in each spectrum. The numbering begins at Val21. High, low, and negative values indicate low, medium, and high flexibility, respectively.
Fig. 3.
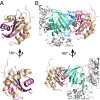
LpoB interacts with the UB2H domain of PBP1B. (A) Residues showing a significant perturbation upon His-UB2H addition are mapped on the cartoon representation of the LpoB(sol) structure. Residues showing chemical shift perturbations higher than 0.05 ppm are shown in pink, and residues for which the [1H,15N] correlation showed intensity changes upon interaction are in dark purple (
SI Appendix, Fig. S4_C_
). (B) Final HADDOCK model of the LpoB–PBP1B complex after addition of ambiguous restraints from NMR perturbations, and biochemical/functional data (Fig. 4). Residues changed to Ala, or reverse-charge amino acids in the activity assays, are represented as sticks in blue (UB2H domain) or magenta (LpoB). Cartoon representation of LpoB and UB2H domain are shown in wheat and green, respectively. Additional domains in PBP1B are shown in gray.
Fig. 4.
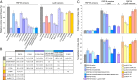
Dissecting the LpoB–PBP1B interaction and its relevance for PBP1B function. (A) In vivo activity of PBP1B and LpoB versions as measured by cellular fitness under cefsulodin treatment (12 and 24 µg/mL for PBP1B and LpoB versions, respectively). Cefsulodin targets primarily PBP1A, increasing the cell’s dependence on PBP1B. Colony size is used as proxy of cellular fitness, and the colony size relative to cells expressing the WT protein is plotted here (mean ± SD; n > 12). (B) _K_d values in μM determined by SPR of the PBP1B–LpoB interaction by using different PBP1B and LpoB versions (mean ± SD, n = 3). n.b., insufficient binding for determination of _K_d; -, not tested. (C) GTase or TPase activity assays for PBP1B using different PBP1B and LpoB versions. The GTase rate is compared with the mean rate of PBP1B alone (mean ± SD; n = 3–6). The TPase activity is shown as percentages of cross-linked peptides in PG (mean ± SD; n = 3–4).
Fig. 5.
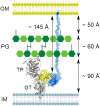
Model of the stimulation of PBP1B by LpoB in the bacterial envelope. LpoB (blue) uses its ∼145-Å-long, flexible N-terminal region to span the periplasm and places its globular domain in position to interact with the UB2H domain of PBP1B (yellow). The thickness of the PG layer and distances to the IM and OM are according to previous studies (19).
Similar articles
- Induced conformational changes activate the peptidoglycan synthase PBP1B.
Egan AJF, Maya-Martinez R, Ayala I, Bougault CM, Banzhaf M, Breukink E, Vollmer W, Simorre JP. Egan AJF, et al. Mol Microbiol. 2018 Nov;110(3):335-356. doi: 10.1111/mmi.14082. Epub 2018 Oct 25. Mol Microbiol. 2018. PMID: 30044025 Free PMC article. - Regulation of Bacterial Peptidoglycan Polymerization.
Arthur M. Arthur M. Trends Microbiol. 2016 Jul;24(7):519-521. doi: 10.1016/j.tim.2016.05.003. Epub 2016 May 25. Trends Microbiol. 2016. PMID: 27236859 - In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli.
Bertsche U, Breukink E, Kast T, Vollmer W. Bertsche U, et al. J Biol Chem. 2005 Nov 11;280(45):38096-101. doi: 10.1074/jbc.M508646200. Epub 2005 Sep 9. J Biol Chem. 2005. PMID: 16154998 - Activities and regulation of peptidoglycan synthases.
Egan AJ, Biboy J, van't Veer I, Breukink E, Vollmer W. Egan AJ, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150031. doi: 10.1098/rstb.2015.0031. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370943 Free PMC article. Review. - Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins).
Pratt RF. Pratt RF. Cell Mol Life Sci. 2008 Jul;65(14):2138-55. doi: 10.1007/s00018-008-7591-7. Cell Mol Life Sci. 2008. PMID: 18408890 Free PMC article. Review.
Cited by
- Transcriptional Responses of Pseudomonas aeruginosa to Inhibition of Lipoprotein Transport by a Small Molecule Inhibitor.
Lorenz C, Dougherty TJ, Lory S. Lorenz C, et al. J Bacteriol. 2020 Nov 19;202(24):e00452-20. doi: 10.1128/JB.00452-20. Print 2020 Nov 19. J Bacteriol. 2020. PMID: 32989085 Free PMC article. - Subunit Arrangement in GpsB, a Regulator of Cell Wall Biosynthesis.
Cleverley RM, Rismondo J, Lockhart-Cairns MP, Van Bentum PT, Egan AJ, Vollmer W, Halbedel S, Baldock C, Breukink E, Lewis RJ. Cleverley RM, et al. Microb Drug Resist. 2016 Sep;22(6):446-60. doi: 10.1089/mdr.2016.0050. Epub 2016 Jun 3. Microb Drug Resist. 2016. PMID: 27257764 Free PMC article. - Conserved mechanism of cell-wall synthase regulation revealed by the identification of a new PBP activator in Pseudomonas aeruginosa.
Greene NG, Fumeaux C, Bernhardt TG. Greene NG, et al. Proc Natl Acad Sci U S A. 2018 Mar 20;115(12):3150-3155. doi: 10.1073/pnas.1717925115. Epub 2018 Mar 5. Proc Natl Acad Sci U S A. 2018. PMID: 29507210 Free PMC article. - Disruption of KPC-producing Klebsiella pneumoniae membrane via induction of oxidative stress by cinnamon bark (Cinnamomum verum J. Presl) essential oil.
Yang SK, Yusoff K, Ajat M, Thomas W, Abushelaibi A, Akseer R, Lim SE, Lai KS. Yang SK, et al. PLoS One. 2019 Apr 2;14(4):e0214326. doi: 10.1371/journal.pone.0214326. eCollection 2019. PLoS One. 2019. PMID: 30939149 Free PMC article. - Plasticity of Escherichia coli cell wall metabolism promotes fitness and antibiotic resistance across environmental conditions.
Mueller EA, Egan AJ, Breukink E, Vollmer W, Levin PA. Mueller EA, et al. Elife. 2019 Apr 9;8:e40754. doi: 10.7554/eLife.40754. Elife. 2019. PMID: 30963998 Free PMC article.
References
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous