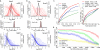Bayesian approach to single-cell differential expression analysis - PubMed (original) (raw)
Bayesian approach to single-cell differential expression analysis
Peter V Kharchenko et al. Nat Methods. 2014 Jul.
Abstract
Single-cell data provide a means to dissect the composition of complex tissues and specialized cellular environments. However, the analysis of such measurements is complicated by high levels of technical noise and intrinsic biological variability. We describe a probabilistic model of expression-magnitude distortions typical of single-cell RNA-sequencing measurements, which enables detection of differential expression signatures and identification of subpopulations of cells in a way that is more tolerant of noise.
Figures
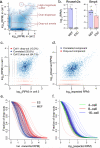
Figure 1. Modeling single-cell RNA-seq measurement as a mixture of two processes
a. Types of cell-to-cell variability observed in single-cell RNA-seq measurements. A smoothed scatter plot compares gene expression estimates from two cells of the same type (MEF cells), illustrating prevalence of drop-out events, over-dispersion, and high-magnitude outliers. b. Single-cell variability throws off standard RNA-seq analysis methods, with top differentially expressed genes influenced by difference in drop-out (Rnaseh2a) or outlier (Bmp4) events. The examples are taken from CuffDiff2 comparison of 10 ESC and 10 MEF cells, with triangles showing expression magnitudes observed in different cells, and whiskers spanning the range of observed expression magnitudes. c. To identify a reliable set of genes for fitting model parameters, our approach initially uses cross-comparison of single-cell measurements (using cells of the same type, e.g. MEF), determining whether the transcript is likely to have been successfully amplified in both experiments (correlated component). The true expression magnitude of such genes is estimated as a median expression level across cells in which the gene appears in a correlated component. d. Each single-cell measurement is modeled as a mixture of drop-out and successful amplification processes. The parameters of the distributions and the magnitude-dependent mixing of the two processes are determined based on the expected population expression averages of genes appearing in many correlated components (c.). e. Drop-out rates vary between different cell types. The rate of transcript detection failures (drop-out events) depends on the average expression magnitude of a gene in the cell population, and varies among the cells. In Islam et al. dataset, higher drop-out frequencies are observed for mouse ES cells compared to MEF cells. f. Drop-out rates for 4, 8 and 16-cell embryo samples examined by Deng et al. using a recently-developed protocol also show systematic differences.
Figure 2. Applying single-cell models for differential expression and subpopulation analyses
a. The model fitted for each single cell is used to estimate the likelihood that a gene is expressed at any particular level (i.e. posterior distribution) given the observed data (colored curves). The approach estimates joint posterior distribution for the overall level with each cell type (black curves), and the expression fold difference between the cell types (middle plot). The example demonstrates expression differences of Sox2 between all ES and MEF cells measured by Islam et al. The plots show posterior probability of expression magnitudes in proximal (top) and distal (bottom) cells. The posterior probability of the fold-expression difference magnitude is shown in the middle plot with the associated raw P-value of differential expression. b. Differential expression of Dazl between cells of 8-cell and 16-cell mouse embryo stages, as determined by SCDE method. A regulator factor expressed in mammalian embryos, , Dazl is expressed at earlier stages, and shows a drop-off between 8- and 16-cell stages. c. The ability of different analysis methods to detect differentially expressed genes is shown using the false/true positive rate relationship (ROC curve), using traditional bulk expression measurements as a benchmark. The SCDE method shows higher sensitivity at low false-positive range, as well as higher overall performance, as measured by area under the curve (AUC) scores. d. Performance of error-model-based transcriptional similarity measures in distinguishing ES and MEF cell types. The plot shows the fraction of correctly classified cells, assessed for increasingly difficult classification problem by iteratively excluding up to 7000 most informative genes (i.e. genes differentially expressed between ES and MEF, x-axis). The 95% confidence bands are shown in light shading. Transcriptional similarity measures that take into account direct or reciprocal drop-out event probability show consistently better classification performance than Pearson linear correlation or Bray-Curtis similarity measure.
Similar articles
- Accounting for technical noise in differential expression analysis of single-cell RNA sequencing data.
Jia C, Hu Y, Kelly D, Kim J, Li M, Zhang NR. Jia C, et al. Nucleic Acids Res. 2017 Nov 2;45(19):10978-10988. doi: 10.1093/nar/gkx754. Nucleic Acids Res. 2017. PMID: 29036714 Free PMC article. - Beyond comparisons of means: understanding changes in gene expression at the single-cell level.
Vallejos CA, Richardson S, Marioni JC. Vallejos CA, et al. Genome Biol. 2016 Apr 15;17:70. doi: 10.1186/s13059-016-0930-3. Genome Biol. 2016. PMID: 27083558 Free PMC article. - Quality Control of Single-Cell RNA-seq.
Jiang P. Jiang P. Methods Mol Biol. 2019;1935:1-9. doi: 10.1007/978-1-4939-9057-3_1. Methods Mol Biol. 2019. PMID: 30758816 - Single-cell RNA-seq: advances and future challenges.
Saliba AE, Westermann AJ, Gorski SA, Vogel J. Saliba AE, et al. Nucleic Acids Res. 2014 Aug;42(14):8845-60. doi: 10.1093/nar/gku555. Epub 2014 Jul 22. Nucleic Acids Res. 2014. PMID: 25053837 Free PMC article. Review. - Normalization for Single-Cell RNA-Seq Data Analysis.
Bacher R. Bacher R. Methods Mol Biol. 2019;1935:11-23. doi: 10.1007/978-1-4939-9057-3_2. Methods Mol Biol. 2019. PMID: 30758817 Review.
Cited by
- A Novel Method to Identify the Differences Between Two Single Cell Groups at Single Gene, Gene Pair, and Gene Module Levels.
Cui L, Wang B, Ren C, Wang A, An H, Liang W. Cui L, et al. Front Genet. 2021 Mar 15;12:648898. doi: 10.3389/fgene.2021.648898. eCollection 2021. Front Genet. 2021. PMID: 33790951 Free PMC article. - Pathway analysis through mutual information.
Jeuken GS, Käll L. Jeuken GS, et al. Bioinformatics. 2024 Jan 2;40(1):btad776. doi: 10.1093/bioinformatics/btad776. Bioinformatics. 2024. PMID: 38195928 Free PMC article. - PD-L1-expressing tumor-associated macrophages are immunostimulatory and associate with good clinical outcome in human breast cancer.
Wang L, Guo W, Guo Z, Yu J, Tan J, Simons DL, Hu K, Liu X, Zhou Q, Zheng Y, Colt EA, Yim J, Waisman J, Lee PP. Wang L, et al. Cell Rep Med. 2024 Feb 20;5(2):101420. doi: 10.1016/j.xcrm.2024.101420. Cell Rep Med. 2024. PMID: 38382468 Free PMC article. - SwarnSeq: An improved statistical approach for differential expression analysis of single-cell RNA-seq data.
Das S, Rai SN. Das S, et al. Genomics. 2021 May;113(3):1308-1324. doi: 10.1016/j.ygeno.2021.02.014. Epub 2021 Mar 1. Genomics. 2021. PMID: 33662531 Free PMC article. - scMTD: a statistical multidimensional imputation method for single-cell RNA-seq data leveraging transcriptome dynamic information.
Qi J, Sheng Q, Zhou Y, Hua J, Xiao S, Jin S. Qi J, et al. Cell Biosci. 2022 Sep 2;12(1):142. doi: 10.1186/s13578-022-00886-4. Cell Biosci. 2022. PMID: 36056412 Free PMC article.
References
- Tang F, et al. Nat Methods. 2009;6:377–382. - PubMed
- Hashimshony T, Wagner F, Sher N, Yanai I. Cell Rep. 2012;2:666–673. - PubMed
Publication types
MeSH terms
Grants and funding
- K25 AG037596/AG/NIA NIH HHS/United States
- U01 HL100402/HL/NHLBI NIH HHS/United States
- R01DK050234-15A1/DK/NIDDK NIH HHS/United States
- R01 HL097794/HL/NHLBI NIH HHS/United States
- K25AG037596/AG/NIA NIH HHS/United States
- R01 DK050234/DK/NIDDK NIH HHS/United States
- R01HL097794-03/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases