Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins - PubMed (original) (raw)
Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins
Yu-Chan Chen et al. EMBO J. 2014.
Abstract
The majority of ER-targeted tail-anchored (TA) proteins are inserted into membranes by the Guided Entry of Tail-anchored protein (GET) system. Disruption of this system causes a subset of TA proteins to mislocalize to mitochondria. We show that the AAA+ ATPase Msp1 limits the accumulation of mislocalized TA proteins on mitochondria. Deletion of MSP1 causes the Pex15 and Gos1 TA proteins to accumulate on mitochondria when the GET system is impaired. Likely as a result of failing to extract mislocalized TA proteins, yeast with combined mutation of the MSP1 gene and the GET system exhibit strong synergistic growth defects and severe mitochondrial damage, including loss of mitochondrial DNA and protein and aberrant mitochondrial morphology. Like yeast Msp1, human ATAD1 limits the mitochondrial mislocalization of PEX26 and GOS28, orthologs of Pex15 and Gos1, respectively. GOS28 protein level is also increased in ATAD1(-/-) mouse tissues. Therefore, we propose that yeast Msp1 and mammalian ATAD1 are conserved members of the mitochondrial protein quality control system that might promote the extraction and degradation of mislocalized TA proteins to maintain mitochondrial integrity.
Keywords: AAA+ ATPase; Guided Entry of Tail‐anchored protein; mitochondrial protein quality control; tail‐anchored proteins.
© 2014 The Authors.
Figures
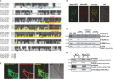
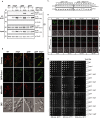
Figure 1. Msp1 is a conserved mitochondrial outer membrane protein
- Protein sequence alignment and domain prediction of yeast, fly, mouse, and human homologs (SDSC Biology Workbench). The transmembrane domain (TMD) is marked by a dashed cyan box (dashed because the endpoints of the TMD are not clearly defined), and the AAA+ domain is marked by a yellow box. Glutamate 193 is indicated by an asterisk (*).
- Representative images of the _msp1_Δ mutant expressing Msp1-GFP and Mito-RFP. Yeast were grown to early log phase at 30°C in synthetic glucose (SD) medium and visualized by fluorescence microscopy. Peroxisomal Msp1-GFP signal is marked by arrows.
- Intact, hypotonically swollen and Triton X-100-solubilized mitochondria of the _msp1_Δ mutant expressing Msp1-His6/HA3 were treated with (+) or without (−) Proteinase K and analyzed by immunoblot with whole-cell lysate (WCL) and post-mitochondrial supernatant (PMS). Mge1, Cyb2, and Fzo1 are matrix, intermembrane space (IMS), and outer membrane proteins (OMM), respectively.
- Soluble (S) and pellet fraction (P) of the KCl- or Na2CO3-treated mitochondria purified from the strain described in (C) were analyzed by immunoblot. Cyb2 is a peripherally attached mitochondrial inner membrane protein; Por1 and Om45 are both integral outer membrane proteins. The asterisk (*) indicates a non-specific band.
- Human dermal fibroblasts (HDFs) stably expressing ATAD1-GFP were stained with Mitotracker Red and visualized by florescence microscopy.
Source data are available online for this figure.
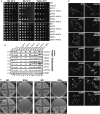
Figure 2. The _msp1_Δ mutant exhibits growth defects and severe mitochondria damage when combined with the GET mutants
- Five-fold dilutions of the indicated yeast strains grown in SD medium were spotted on SD or synthetic glycerol plates (SGly) and incubated at 30 or 37°C.
- Whole-cell lysates of log-phase cultures harvested from synthetic raffinose medium were analyzed by immunoblot. Both mitochondrial and nuclear DNA-encoded mitochondrial proteins were immunoblotted. Pgk1 is a cytoplasmic control protein.
- The parental haploids and two independent diploids were streaked on SD and SGly plates and cultured at 30°C. The _sdh5_Δ strain is a control representing a respiratory deficient rho + strain.
- Representative images of the indicated yeast strains expressing the mito-RFP construct, grown in SD medium to log phase and visualized by fluorescence microscopy. Areas of atypical mitochondrial swelling are indicated by arrows (see also Supplementary Fig S2E). The RFP image intensity was adjusted to be similar for visualization purposes. Unadjusted images from similar exposure times are shown in Supplementary Fig S2F.
Source data are available online for this figure.
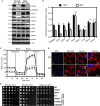
Figure 3. Depletion of ATAD1 causes decreased mitochondrial protein level and mitochondrial fragmentation in mammals
- Immunoblots of whole-brain lysates obtained from the wild-type (WT) and _ATAD1_−/− knockout (KO) mice. PDH, pyruvate dehydrogenase; HXK1 and 2, hexokinase 1 and 2.
- The optical densitometry quantification of (A). The values represent the mean ± SEM (n = 3, *P < 0.05, **P < 0.005, ***P < 0.001, one-way ANOVA, Tukey's multiple comparison tests).
- Oxygen consumption rate (OCR) of the WT and _ATAD1_−/− mouse embryonic fibroblasts (MEFs). The data were normalized to the amount of protein in each well and represent % control. **P < 0.005, ***P < 0.001.
- Representative images of the mitochondrial morphology of the WT and _ATAD1_−/− MEFs. The cells were transfected with Mito-RFP (red), stained with DAPI (blue), and visualized.
- Five-fold dilutions of the indicated strains harboring empty vector (EV), yeast Msp1, human ATAD1 or fly CG5395 construct cultured overnight in SD-Ura medium were spotted on SD-Ura or SGly-Ura plates and incubated at 30 or 37°C.
Source data are available online for this figure.
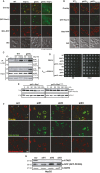
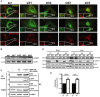
Figure 4. Yeast Msp1 and human ATAD1 are required to limit the level of mitochondrially mislocalized TA proteins Pex15 and PEX26, respectively
- Representative images of GFP-Pex15 localization. The indicated yeast strains expressing GFP-Pex15 from the GAL1 promoter and plasmid-borne Mito-RFP were grown overnight in SD medium, switched to galactose induction medium for 6 h, and visualized by fluorescence microscopy. The _get3_Δ _msp1_Δ images (1,000 ms) were exposed 5 times longer than the WT,_msp1_Δ, and _get3_Δ images (200 ms).
- The WT and _get3_Δ strain expressing GFP-Pex15 from the GAL1 promoter and the plasmid-borne Mito-RFP were transformed with either EV or a Msp1 overexpressing construct (Msp1(2μ)) and subjected to the same procedure as described in (A).
- Pex15 co-immunoprecipitates with the ‘trap mutant’ Msp1E193Q. Msp1 was immunoprecipitated by anti-HA antibodies from digitonin-solubilized mitochondria that were extracted from the indicated strains overexpressing GFP-Pex15 from the GAL1 promoter and empty vector (−), wild-type Msp1 (WT) or Msp1E193Q (E193Q). Pex15 expression was induced in galactose medium for 6 h. 4% of the crude lysates, and final eluates were immunoblotted with anti-HA and GFP. Om45 is a mitochondrial outer membrane protein and is used as a loading control.
- Five-fold dilutions of the indicated strains expressing Pex15 from the native PEX15 promoter or GAL1 promoter (P GAL 1::GFP–PEX15) were grown overnight in SD medium, spotted on SD or synthetic galactose (SGal) plates, and cultured at 30°C.
- The _get3_Δ strain co-expressing GFP-Pex15 from the GAL1 promoter and wild-type Msp1 or Msp1E193Q was pulsed in galactose medium for 5 h to induce GFP-Pex15 accumulation and chased in glucose medium to shut off transcription. Whole-cell lysates were prepared from cells harvested every 45 min and analyzed by immunoblot. Por1 is the loading control.
- HepG2 cells stably expressing GFP-PEX26 were treated with scrambled siRNA (scr) or siRNAs (#1–3) targeting human ATAD1 for 6 days, stained with Mitotracker Red, and visualized by fluorescence microscopy with equivalent exposure times.
- Whole-cell lysates of cells from (F) were analyzed by immunoblot using the indicated antibodies. QCR2 is a mitochondrial protein and is used as a loading control.
Source data are available online for this figure.
Figure 5. Msp1 physically interacts with the TA protein Gos1 and is required to limit mitochondrial Gos1
- Gos1 co-immunoprecipitates with the ‘trap mutant’ Msp1. Msp1 was immunoprecipitated by anti-HA antibody from digotonin-solubilized mitochondria that were extracted from the indicated strains expressing GFP-Gos1 from the native GOS1 promoter and empty vector (−), wild-type Msp1 (+) or Msp1E193Q (E193Q). 4% of the crude lysates and final eluates were immunoblotted with anti-HA, GFP, and Om45 antibodies. Om45 is a control mitochondrial outer membrane protein.
- The indicated strains expressing GFP-Gos1 from the native promoter and Mito-RFP were grown in SD medium to early log phase and visualized by fluorescence microscopy.
- The _get3_Δ strain expressing GFP-Gos1 from the GAL1 promoter was transformed with EV or Msp1E193Q, grown in galactose medium for 5 h to induce Gos1 expression and then switched to glucose medium to shut off transcription. Cells were harvested every 30 min thereafter, and the whole-cell lysates were analyzed by immunoblot.
- Representative images of the GAL1 promoter-based transcriptional shut-off experiment as described in (C) except that the indicated strains were co-transformed with Mito-RFP to visualize mitochondria.
- Five-fold dilutions of the indicated strains harboring either empty vector (−) or Gos1 overexpression 2 μ vector (+) were spotted on SD or SGly plates and incubated at 30 or 37°C.
Source data are available online for this figure.
Figure 6. ATAD1 physically interacts with the TA protein, GOS28, and is required to limit the level of mislocalized GOS28 on mitochondria in mammals
- Human dermal fibroblasts (HDFs) stably expressing GFP-GOS28 were treated with scr or siRNA (#1-4) against h_ATAD_ 1, stained with Mitotracker Red, and visualized by fluorescence microscopy.
- Whole-cell lysate of cells from (A) were immunoblotted using anti-ATAD1 and actin (loading control) antibodies.
- Crude mitochondria were extracted from HepG2 cells that stably co-express GFP-GOS28 with empty vector (−), HA-tagged wild-type ATAD1 (WT), or ATAD1E193Q mutant. ATAD1 was immunoprecipitated using anti-HA antibody from digitonin-solubilized lysates and 5% of the crude lysates, and eluates were immunoblotted with anti-GFP and HA antibodies.
- Mouse tissue lysates (30 μg protein) from three WT and _ATAD1_−/− mice were analyzed by immunoblot. S6 ribosomal protein and actin are used as loading controls.
- The optical densitometry quantification of (D). The values represent the mean ± SEM (***P < 0.001, one-way ANOVA).
Source data are available online for this figure.
Comment in
- Msp1: patrolling mitochondria for lost proteins.
Hegde RS. Hegde RS. EMBO J. 2014 Jul 17;33(14):1509-10. doi: 10.15252/embj.201488930. Epub 2014 Jun 10. EMBO J. 2014. PMID: 24916308 Free PMC article.
Similar articles
- The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins.
Okreglak V, Walter P. Okreglak V, et al. Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):8019-24. doi: 10.1073/pnas.1405755111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821790 Free PMC article. - Mitochondrial AAA-ATPase Msp1 detects mislocalized tail-anchored proteins through a dual-recognition mechanism.
Li L, Zheng J, Wu X, Jiang H. Li L, et al. EMBO Rep. 2019 Apr;20(4):e46989. doi: 10.15252/embr.201846989. Epub 2019 Mar 11. EMBO Rep. 2019. PMID: 30858337 Free PMC article. - Msp1 Is a Membrane Protein Dislocase for Tail-Anchored Proteins.
Wohlever ML, Mateja A, McGilvray PT, Day KJ, Keenan RJ. Wohlever ML, et al. Mol Cell. 2017 Jul 20;67(2):194-202.e6. doi: 10.1016/j.molcel.2017.06.019. Epub 2017 Jul 14. Mol Cell. 2017. PMID: 28712723 Free PMC article. - Quality control pathways of tail-anchored proteins.
Jiang H. Jiang H. Biochim Biophys Acta Mol Cell Res. 2021 Feb;1868(2):118922. doi: 10.1016/j.bbamcr.2020.118922. Epub 2020 Dec 4. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33285177 Review. - Msp1/ATAD1 in Protein Quality Control and Regulation of Synaptic Activities.
Wang L, Walter P. Wang L, et al. Annu Rev Cell Dev Biol. 2020 Oct 6;36:141-164. doi: 10.1146/annurev-cellbio-031220-015840. Epub 2020 Sep 4. Annu Rev Cell Dev Biol. 2020. PMID: 32886535 Review.
Cited by
- GET pathway mediates transfer of mislocalized tail-anchored proteins from mitochondria to the ER.
Matsumoto S, Ono S, Shinoda S, Kakuta C, Okada S, Ito T, Numata T, Endo T. Matsumoto S, et al. J Cell Biol. 2022 Jun 6;221(6):e202104076. doi: 10.1083/jcb.202104076. Epub 2022 Apr 20. J Cell Biol. 2022. PMID: 35442388 Free PMC article. - A selectivity filter in the ER membrane protein complex limits protein misinsertion at the ER.
Pleiner T, Hazu M, Pinton Tomaleri G, Nguyen VN, Januszyk K, Voorhees RM. Pleiner T, et al. J Cell Biol. 2023 Aug 7;222(8):e202212007. doi: 10.1083/jcb.202212007. Epub 2023 May 18. J Cell Biol. 2023. PMID: 37199759 Free PMC article. - Mitochondria-Associated Degradation Pathway (MAD) Function beyond the Outer Membrane.
Liao PC, Wolken DMA, Serrano E, Srivastava P, Pon LA. Liao PC, et al. Cell Rep. 2020 Jul 14;32(2):107902. doi: 10.1016/j.celrep.2020.107902. Cell Rep. 2020. PMID: 32668258 Free PMC article. - The GET pathway serves to activate Atg32-mediated mitophagy by ER targeting of the Ppg1-Far complex.
Onishi M, Kubota M, Duan L, Tian Y, Okamoto K. Onishi M, et al. Life Sci Alliance. 2023 Jan 25;6(4):e202201640. doi: 10.26508/lsa.202201640. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36697253 Free PMC article. - A trap mutant reveals the physiological client spectrum of TRC40.
Coy-Vergara J, Rivera-Monroy J, Urlaub H, Lenz C, Schwappach B. Coy-Vergara J, et al. J Cell Sci. 2019 Jul 1;132(13):jcs230094. doi: 10.1242/jcs.230094. J Cell Sci. 2019. PMID: 31182645 Free PMC article.
References
- Boldogh IR, Pon LA. Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2007;80:45–64. - PubMed
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, Redin C, Boudina S, Gygi SP, Brivet M, Thummel CS, Rutter J. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG029368/AG/NIA NIH HHS/United States
- 5R01AG029368/AG/NIA NIH HHS/United States
- P50 DA000266/DA/NIDA NIH HHS/United States
- R01 GM094232/GM/NIGMS NIH HHS/United States
- DA000266/DA/NIDA NIH HHS/United States
- R01GM094232/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases