The oligomeric states of the purified sigma-1 receptor are stabilized by ligands - PubMed (original) (raw)
The oligomeric states of the purified sigma-1 receptor are stabilized by ligands
Katarzyna A Gromek et al. J Biol Chem. 2014.
Abstract
Sigma-1 receptor (S1R) is a mammalian member of the ERG2 and sigma-1 receptor-like protein family (pfam04622). It has been implicated in drug addiction and many human neurological disorders, including Alzheimer and Parkinson diseases and amyotrophic lateral sclerosis. A broad range of synthetic small molecules, including cocaine, (+)-pentazocine, haloperidol, and small endogenous molecules such as N,N-dimethyltryptamine, sphingosine, and steroids, have been identified as regulators of S1R. However, the mechanism of activation of S1R remains obscure. Here, we provide evidence in vitro that S1R has ligand binding activity only in an oligomeric state. The oligomeric state is prone to decay into an apparent monomeric form when exposed to elevated temperature, with loss of ligand binding activity. This decay is suppressed in the presence of the known S1R ligands such as haloperidol, BD-1047, and sphingosine. S1R has a GXXXG motif in its second transmembrane region, and these motifs are often involved in oligomerization of membrane proteins. Disrupting mutations within the GXXXG motif shifted the fraction of the higher oligomeric states toward smaller states and resulted in a significant decrease in specific (+)-[(3)H]pentazocine binding. Results presented here support the proposal that S1R function may be regulated by its oligomeric state. Possible mechanisms of molecular regulation of interacting protein partners by S1R in the presence of small molecule ligands are discussed.
Keywords: GXXXG Motif; Ligand-binding Protein; Membrane Protein; Protein-Protein Interaction; Signal Transduction; σ Receptor.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
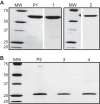
FIGURE 1.
SDS-PAGE of MBP-4A-S1R (A) and S1R (B). The starting MBP-4A-S1R and S1R preparations prior to SEC separation are labeled P1 and P2, respectively. Peaks 1–4 from Fig. 2_A_ are labeled respectively. Molecular mass markers (MW) are labeled in kDa.
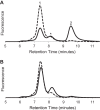
FIGURE 2.
Evidence from size exclusion chromatography for oligomeric states of S1R. A, elution profiles for MBP-4A-S1R (solid line) and S1R (dashed line). Peaks corresponding to different oligomerization states are marked as 1–4. B, repeat chromatography of peaks 1 (solid line) and 3 (dashed line) from A. C, repeat chromatography of peaks 2 (solid line) and 4 (dashed line) from A.
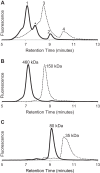
FIGURE 3.
Oligomeric molecular weight of MBP-4A-S1R determined by light scattering. Elution profiles detected by 652-nm laser light scattering are shown by dashed lines. Oligomeric stoichiometry across each peak is marked with a solid line. Peaks have the same labeling as in Fig. 1 and 2. A, apparent monomer is confirmed to be a monomer. B, largest molecular weight oligomer is polydisperse, with stoichiometry ranging from a hexamer to octamer. C, intermediate oligomer is a tetramer.
FIGURE 4.
Analysis of oligomeric states of MBP-4A-S1R by chemical cross-linking agent DSS. DSS-free controls are shown by lanes 1 and 2, containing 0 and 10 μ
m
high affinity ligand BD-1047, respectively. Lanes 3 and 4 mirror the control with the addition of DSS. A, addition of cross-linking agent to the monomer (Fig. 2_A, peak 2_). The bands show slight smearing with no shift in size, signifying only intramolecular cross-linking. B, addition of cross-linking agent to the oligomer (Fig. 2_A_, peak 1). The bands show a mass greater than 300 kDa, and oligomeric stoichiometry cannot be accurately determined. However, an oligomeric state greater than tetramer is clearly visible. C, addition of cross-linking agent to the intermediate oligomer (Fig. 2_A, peak_ *). The bands show an approximate 4-fold increase in size to a mass between 250 and 300 kDa, suggesting a tetrameric state.
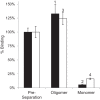
FIGURE 5.
Comparison of specific pentazocine binding activity of S1R oligomers before and after repeat chromatography of peaks from Fig. 2, B and C. The black bars are for assays of MBP-4A-S1R; white bars are for assays of S1R. Binding assays were performed in triplicate, and the error bars represent 1σ deviation.
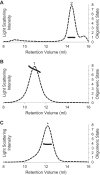
FIGURE 6.
Stabilization of oligomeric S1R by ligand binding. A, size exclusion chromatogram of MBP-4A-S1R before (dashed line) and after incubation for 1 day at 37 °C (solid line) without added ligand. Peaks have the same labeling as in Fig. 1 and 2. B, chromatogram of MBP-4A-S1R before (dashed line) and after (solid line) incubation for 1 day at 37 °C in the presence of the tight binding ligand BD-1047 (0.45 μ
m
).
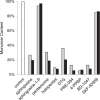
FIGURE 7.
Comparison of the ability of various S1R ligands to prevent conversion to the inactive monomer state. The amount of MBP-4A-S1R converted to the monomer (peak 2 from Fig. 2) in the absence of ligands served as the control (white bar). Gray and black bars indicate ligand doses of 0.45 and 10 μ
m
, respectively, whereas the concentration of MBP-4A-S1R was always 0.23 μ
m
. Tight-binding ligands 4-PPBP, BD-1047 and others stabilized the oligomeric states, whereas sphingosine 1-phosphate allowed conversion to the monomeric state.
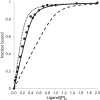
FIGURE 8.
Ligand binding stoichiometry determined by titration of BD-1047 into a 300 nm sample of peak 1 (see Fig. 2_B_). Experimental measurements (solid circles) were made in the concentration range from 0 to 3000 n
m
, with results shown up to 600 n
m
ligand. Binding curves were calculated as described under “Experimental Procedures” with best fit values of KD = 7 n
m
and n = 0.43 (solid line), fixed KD = 7 n
m
and n = 0.25 (dotted line), or fixed KD = 7 n
m
and n = 1 (dashed line).
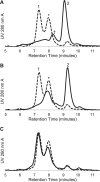
FIGURE 9.
Size exclusion chromatography of MBP-4A-S1R with mutations in the G_XXX_G motif. The control chromatogram of MBP-4A-S1R lacking mutations is shown as a dotted line. A, G91I MBP-4A-S1R with defined peaks as in Fig. 2. A significant shift toward the monomeric state is seen, as is a new ∼180 kDa peak labeled with †. B, G91L MBP-4A-S1R showing conversion to intermediate oligomeric (*) and monomer (2nd peak) states. C, H97A MBP-4A-S1R showed little change in the oligomerization states relative to the nonmutated receptor.
FIGURE 10.
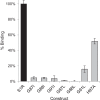
[3H]Pentazocine-specific binding for MBP-4A-S1R and the variants with mutations in the G_XXX_G motif. Binding activity is shown relative to MBP-4A-S1R (black bar); n = 3; error bars represent 1σ deviation.
FIGURE 11.
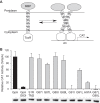
TOXCAT measurements for mutations of the TM2 domain of S1R. A, schematic of the TOXCAT experiment, where periplasmic secretion of MBP is used to place a TM domain into the cytoplasmic membrane, whereas ToxR resides in the cytoplasm. Dimerization of the TM promotes dimerization of ToxR, which then binds to the ctx promoter acting as a transcriptional activator, in this case for CAT. B, TOXCAT response is reported as a percentage relative to the strong transmembrane oligomerization control, glycophorin A (GpA). Immunoblot results obtained from anti-MBP-HRP are shown below the histogram bars and indicate equivalent expression of all TM2 domain variants.
FIGURE 12.
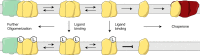
Model representing the proposed interconversions of S1R between the monomer form and ligand-stabilized oligomeric forms. Protein partners of the monomer form would include voltage-gated Na+ channel, acid-sensing channels, and dopamine D1 receptor (16, 23, 73).
Similar articles
- The sigma-1 receptors are present in monomeric and oligomeric forms in living cells in the presence and absence of ligands.
Mishra AK, Mavlyutov T, Singh DR, Biener G, Yang J, Oliver JA, Ruoho A, Raicu V. Mishra AK, et al. Biochem J. 2015 Mar 1;466(2):263-271. doi: 10.1042/BJ20141321. Biochem J. 2015. PMID: 25510962 Free PMC article. - Improved expression and purification of sigma 1 receptor fused to maltose binding protein by alteration of linker sequence.
Gromek KA, Meddaugh HR, Wrobel RL, Suchy FP, Bingman CA, Primm JG, Fox BG. Gromek KA, et al. Protein Expr Purif. 2013 Jun;89(2):203-9. doi: 10.1016/j.pep.2013.03.013. Epub 2013 Apr 3. Protein Expr Purif. 2013. PMID: 23562661 Free PMC article. - Biochemical Pharmacology of the Sigma-1 Receptor.
Chu UB, Ruoho AE. Chu UB, et al. Mol Pharmacol. 2016 Jan;89(1):142-53. doi: 10.1124/mol.115.101170. Epub 2015 Nov 11. Mol Pharmacol. 2016. PMID: 26560551 Review. - Distinct Regulation of σ 1 Receptor Multimerization by Its Agonists and Antagonists in Transfected Cells and Rat Liver Membranes.
Hong WC. Hong WC. J Pharmacol Exp Ther. 2020 May;373(2):290-301. doi: 10.1124/jpet.119.262790. Epub 2020 Feb 14. J Pharmacol Exp Ther. 2020. PMID: 32060048 - A Review of the Human Sigma-1 Receptor Structure.
Ossa F, Schnell JR, Ortega-Roldan JL. Ossa F, et al. Adv Exp Med Biol. 2017;964:15-29. doi: 10.1007/978-3-319-50174-1_3. Adv Exp Med Biol. 2017. PMID: 28315262 Review.
Cited by
- Novel missense alleles of SIGMAR1 as tools to understand emerin-dependent gene silencing in response to cocaine.
Arun AS, Eddings CR, Wilson KL. Arun AS, et al. Exp Biol Med (Maywood). 2019 Nov;244(15):1354-1361. doi: 10.1177/1535370219863444. Epub 2019 Jul 19. Exp Biol Med (Maywood). 2019. PMID: 31324122 Free PMC article. - The sigma-1 receptor behaves as an atypical auxiliary subunit to modulate the functional characteristics of Kv1.2 channels expressed in HEK293 cells.
Abraham MJ, Fleming KL, Raymond S, Wong AYC, Bergeron R. Abraham MJ, et al. Physiol Rep. 2019 Jul;7(12):e14147. doi: 10.14814/phy2.14147. Physiol Rep. 2019. PMID: 31222975 Free PMC article. - Pharmacological profiling of sigma 1 receptor ligands by novel receptor homomer assays.
Yano H, Bonifazi A, Xu M, Guthrie DA, Schneck SN, Abramyan AM, Fant AD, Hong WC, Newman AH, Shi L. Yano H, et al. Neuropharmacology. 2018 May 1;133:264-275. doi: 10.1016/j.neuropharm.2018.01.042. Epub 2018 Jan 31. Neuropharmacology. 2018. PMID: 29407216 Free PMC article. - In vitro and in vivo sigma 1 receptor imaging studies in different disease states.
Agha H, McCurdy CR. Agha H, et al. RSC Med Chem. 2020 Oct 8;12(2):154-177. doi: 10.1039/d0md00186d. eCollection 2021 Mar 4. RSC Med Chem. 2020. PMID: 34046607 Free PMC article. Review.
References
- Jbilo O., Vidal H., Paul R., De Nys N., Bensaid M., Silve S., Carayon P., Davi D., Galiègue S., Bourrié B., Guillemot J. C., Ferrara P., Loison G., Maffrand J. P., Le Fur G., Casellas P. (1997) Purification and characterization of the human SR 31747A-binding protein. A nuclear membrane protein related to yeast sterol isomerase. J. Biol. Chem. 272, 27107–27115 - PubMed
- Kekuda R., Prasad P. D., Fei Y. J., Leibach F. H., Ganapathy V. (1996) Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1). Biochem. Biophys. Res. Commun. 229, 553–558 - PubMed
- Seth P., Leibach F. H., Ganapathy V. (1997) Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem. Biophys. Res. Commun. 241, 535–540 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1S10RR023748-01/RR/NCRR NIH HHS/United States
- R21 NS075820/NS/NINDS NIH HHS/United States
- S10 RR023748/RR/NCRR NIH HHS/United States
- NS075820/NS/NINDS NIH HHS/United States
- U54 GM094584/GM/NIGMS NIH HHS/United States
- R01 GM099752/GM/NIGMS NIH HHS/United States
- R01GM099752/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources