Helminth colonization is associated with increased diversity of the gut microbiota - PubMed (original) (raw)
. 2014 May 22;8(5):e2880.
doi: 10.1371/journal.pntd.0002880. eCollection 2014 May.
Mei San Tang 2, Yvonne A L Lim 1, Seow Huey Choy 1, Zachary D Kurtz 3, Laura M Cox 3, Uma Mahesh Gundra 4, Ilseung Cho 5, Richard Bonneau 6, Martin J Blaser 3, Kek Heng Chua 7, P'ng Loke 4
Affiliations
- PMID: 24851867
- PMCID: PMC4031128
- DOI: 10.1371/journal.pntd.0002880
Helminth colonization is associated with increased diversity of the gut microbiota
Soo Ching Lee et al. PLoS Negl Trop Dis. 2014.
Erratum in
- Correction: Helminth Colonization Is Associated with Increased Diversity of the Gut Microbiota.
Lee SC, Tang MS, Lim YAL, Choy SH, Kurtz ZD, Cox LM, Gundra UM, Cho I, Bonneau R, Blaser MJ, Chua KH, Loke P. Lee SC, et al. PLoS Negl Trop Dis. 2021 Apr 7;15(4):e0009325. doi: 10.1371/journal.pntd.0009325. eCollection 2021 Apr. PLoS Negl Trop Dis. 2021. PMID: 33826625 Free PMC article.
Abstract
Soil-transmitted helminths colonize more than 1.5 billion people worldwide, yet little is known about how they interact with bacterial communities in the gut microbiota. Differences in the gut microbiota between individuals living in developed and developing countries may be partly due to the presence of helminths, since they predominantly infect individuals from developing countries, such as the indigenous communities in Malaysia we examine in this work. We compared the composition and diversity of bacterial communities from the fecal microbiota of 51 people from two villages in Malaysia, of which 36 (70.6%) were infected by helminths. The 16S rRNA V4 region was sequenced at an average of nineteen thousand sequences per samples. Helminth-colonized individuals had greater species richness and number of observed OTUs with enrichment of Paraprevotellaceae, especially with Trichuris infection. We developed a new approach of combining centered log-ratio (clr) transformation for OTU relative abundances with sparse Partial Least Squares Discriminant Analysis (sPLS-DA) to enable more robust predictions of OTU interrelationships. These results suggest that helminths may have an impact on the diversity, bacterial community structure and function of the gut microbiota.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
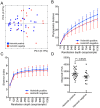
Figure 1. Abundance and diversity in the intestinal microbiome in 51 Malaysian subjects.
(Panel A) The number of observed OTUs plotted against age for all 51 individual samples. The number of observed OTUs for most samples was between 1500–4000. (Panel B) Relative abundance of the top phyla represented across the 51 subjects arranged by increasing age. The abundance patterns were largely similar across the individual subjects, except in two of the younger subjects who had high abundance of Actinobacteria (Bifidobacterium sp.) in their stool samples.
Figure 2. Beta and alpha diversity for the 51 subjects.
(Panel A) PCoA of the microbial communities in helminth-positive and helminth-negative samples. Clustering of helminth-positive subjects could be observed, which is statistically significant (p = 0.04). Rarefaction curves calculated for phylogenetic distance (Panel B) and Shannon index (Panel C) demonstrating the higher microbial diversity found among helminth positive subjects. (Panel D) Total number of observed OTUs in each individual sample was compared between helminth positive and negative groups.
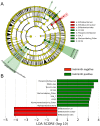
Figure 3. Different abundances of bacterial communities between helminth-positive and negative subjects.
With LEfSe for data analysis and visualization key OTUs were identified as differentiating between helminth-positive and helminth-negative fecal samples. (Panel A) Bacterial taxa that were differentially abundant in the gut microbiota profiles of helminth positive and helminth negative subjects visualized using a cladogram generated from LEfSe analysis. (Panel B) With a log LDA score above 3.00, we found an increased abundance of OTUs contributed by Paraprevotellaceae, Mollicutes, Bacteroidales and Alphaproteobacteria among helminth-positive subjects, while helminth-negative subjects had increased abundance of Bifidobacterium.
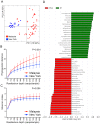
Figure 4. Identification of helminth associated OTUs by centered log-ratio (clr) transformations and sparse Partial Least Squares Discriminant Analysis (sPLS-DA).
Supervised sPLS-DA models were first used to identify OTUs associated with helminth infection (Panel A), Trichuris alone (Panel B) or Ascaris alone (Panel C) with statistically significant model coefficients (alpha = 0.05). These OTUs then underwent additional permutation testing by fitting sPLS-DA models to over a thousand randomized datasets. These models were then visualized using biplots of the first two PLS factors.
Figure 5. Inferred metagenomic analyses with PICRUSt.
(Panel A) Relative abundances of KEGG pathways encoded in the gut microbiota of helminth positive and negative indigenous Malaysians. (Panel B) Supervised comparison using LEfSe identifies differentially abundant KEGG pathways (Log LDA>2.00) in individuals positive for helminths. No functional pathways were differentially abundant in the helminth negative individuals. (Panel C) A similar list of functional pathways was found to be differentially abundant in individuals infected with Trichuris alone. (Panel D) Comparison between individuals who were positive for Ascaris only and individuals who were negative for helminths found translational pathways to be differentially abundant in the former (labeled green) and carbohydrate metabolism pathway to be differentially abundant in the latter group (labeled red). (Panel E) Genes in the pathways of Replication and repair, Translation, Nucleotide metabolism and Cell growth and death are more abundant in helminth positive individuals.
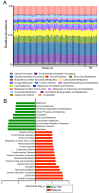
Figure 6. Differences in gut microbiota between New York and Malaysian subjects.
(Panel A) PCoA plot for beta-diversity patterns of bacterial communities of fecal samples from the Malaysian indigenous community and the New Yorkers. Rarefaction curves plotted for phylogenetic distance (Panel B) and Shannon index (Panel C) as measures of alpha diversity. Phylogenetic distance and Shannon index were calculated at a rarefaction depth of 1450sequences/sample, and showed significant differences between the Malaysian and New York samples. (Panel D) LEfSe analysis of fecal communities showing Firmicutes as the differentially abundant phylum in the New York stool samples (green, NY) while Cyanobacteria, Actinobacteria, Tenericutes, and Gammaproteobacteria were differentially abundant in the Malaysian stool samples (red, MSIA).
Figure 7. Metagenomic differences between New York and Malaysian subjects.
(Panel A) Relative abundances of metabolic pathways encoded in the gut microbiota of indigenous Malaysians and New Yorkers. (Panel B) Functional divergence between the gut microbiota of indigenous Malaysians and New Yorkers. Supervised comparison identifies differential abundance of specific KEGG pathways using LEfSe (Log LDA>2.00).
Similar articles
- Helminth-Induced Human Gastrointestinal Dysbiosis: a Systematic Review and Meta-Analysis Reveals Insights into Altered Taxon Diversity and Microbial Gradient Collapse.
Kupritz J, Angelova A, Nutman TB, Gazzinelli-Guimaraes PH. Kupritz J, et al. mBio. 2021 Dec 21;12(6):e0289021. doi: 10.1128/mBio.02890-21. Epub 2021 Dec 21. mBio. 2021. PMID: 34933444 Free PMC article. - Helminths, polyparasitism, and the gut microbiome in the Philippines.
Gordon CA, Krause L, McManus DP, Morrison M, Weerakoon KG, Connor MC, Olveda RM, Ross AG, Gobert GN. Gordon CA, et al. Int J Parasitol. 2020 Mar;50(3):217-225. doi: 10.1016/j.ijpara.2019.12.008. Epub 2020 Mar 3. Int J Parasitol. 2020. PMID: 32135180 - Exposure to Parasitic Protists and Helminths Changes the Intestinal Community Structure of Bacterial Communities in a Cohort of Mother-Child Binomials from a Semirural Setting in Mexico.
Partida-Rodriguez O, Nieves-Ramirez M, Laforest-Lapointe I, Brown EM, Parfrey L, Valadez-Salazar A, Thorson L, Morán P, Gonzalez E, Rascon E, Magaña U, Hernandez E, Rojas-Velázquez L, Torres J, Arrieta MC, Ximenez C, Finlay BB. Partida-Rodriguez O, et al. mSphere. 2021 Aug 25;6(4):e0008321. doi: 10.1128/mSphere.00083-21. Epub 2021 Aug 18. mSphere. 2021. PMID: 34406855 Free PMC article. - Immune Response and Microbiota Profiles during Coinfection with Plasmodium vivax and Soil-Transmitted Helminths.
Easton AV, Raciny-Aleman M, Liu V, Ruan E, Marier C, Heguy A, Yasnot MF, Rodriguez A, Loke P. Easton AV, et al. mBio. 2020 Oct 20;11(5):e01705-20. doi: 10.1128/mBio.01705-20. mBio. 2020. PMID: 33082257 Free PMC article. - Helminths and Bacterial Microbiota: The Interactions of Two of Humans' "Old Friends".
Llinás-Caballero K, Caraballo L. Llinás-Caballero K, et al. Int J Mol Sci. 2022 Nov 1;23(21):13358. doi: 10.3390/ijms232113358. Int J Mol Sci. 2022. PMID: 36362143 Free PMC article. Review.
Cited by
- Sparse and compositionally robust inference of microbial ecological networks.
Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Kurtz ZD, et al. PLoS Comput Biol. 2015 May 7;11(5):e1004226. doi: 10.1371/journal.pcbi.1004226. eCollection 2015 May. PLoS Comput Biol. 2015. PMID: 25950956 Free PMC article. - Increasing helminth infection burden depauperates the diversity of the gut microbiota and alters its composition in mice.
Guiver E, Galan M, Lippens C, Bellenger J, Faivre B, Sorci G. Guiver E, et al. Curr Res Parasitol Vector Borne Dis. 2022 Feb 22;2:100082. doi: 10.1016/j.crpvbd.2022.100082. eCollection 2022. Curr Res Parasitol Vector Borne Dis. 2022. PMID: 36589866 Free PMC article. - The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease.
Zuo T, Ng SC. Zuo T, et al. Front Microbiol. 2018 Sep 25;9:2247. doi: 10.3389/fmicb.2018.02247. eCollection 2018. Front Microbiol. 2018. PMID: 30319571 Free PMC article. Review. - The Underrated Gut Microbiota Helminths, Bacteriophages, Fungi, and Archaea.
Garcia-Bonete MJ, Rajan A, Suriano F, Layunta E. Garcia-Bonete MJ, et al. Life (Basel). 2023 Aug 18;13(8):1765. doi: 10.3390/life13081765. Life (Basel). 2023. PMID: 37629622 Free PMC article. Review. - Assessment of Electronic Sensing Techniques for the Rapid Identification of Alveolar Echinococcosis through Exhaled Breath Analysis.
Kwiatkowski A, Chludziński T, Saidi T, Welearegay TG, Jaimes-Mogollón AL, El Bari N, Borys S, Bouchikhi B, Smulko J, Ionescu R. Kwiatkowski A, et al. Sensors (Basel). 2020 May 7;20(9):2666. doi: 10.3390/s20092666. Sensors (Basel). 2020. PMID: 32392783 Free PMC article.
References
- WHO (2012) Soil-transmitted helminth infections. Fact sheet No 366.
Publication types
MeSH terms
Grants and funding
- R01 AI093811/AI/NIAID NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- AI094166/AI/NIAID NIH HHS/United States
- AI093811/AI/NIAID NIH HHS/United States
- R01 DK090989/DK/NIDDK NIH HHS/United States
- R21 AI094166/AI/NIAID NIH HHS/United States
- T32 AI007180/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources