Large-scale, protection-free synthesis of Se-adenosyl-L-selenomethionine analogues and their application as cofactor surrogates of methyltransferases - PubMed (original) (raw)
. 2014 Jun 6;16(11):3056-9.
doi: 10.1021/ol501169y. Epub 2014 May 22.
Affiliations
- PMID: 24852128
- PMCID: PMC4059250
- DOI: 10.1021/ol501169y
Large-scale, protection-free synthesis of Se-adenosyl-L-selenomethionine analogues and their application as cofactor surrogates of methyltransferases
Ian R Bothwell et al. Org Lett. 2014.
Abstract
S-adenosyl-L-methionine (SAM) analogues have previously demonstrated their utility as chemical reporters of methyltransferases. Here we describe the facile, large-scale synthesis of Se-alkyl Se-adenosyl-L-selenomethionine (SeAM) analogues and their precursor, Se-adenosyl-L-selenohomocysteine (SeAH). Comparison of SeAM analogues with their equivalent SAM analogues suggests that sulfonium-to-selenonium substitution can enhance their compatibility with certain protein methyltransferases, favoring otherwise less reactive SAM analogues. Ready access to SeAH therefore enables further application of SeAM analogues as chemical reporters of diverse methyltransferases.
Figures
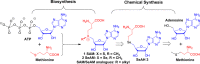
Figure 1
Biosynthesis of SAM and retrosynthesis of SAM, SeAM, and their chalcogen–alkyl analogues.
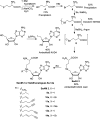
Scheme 1. Protection-Free Synthesis of SeAH, SeAM, and SeAM Analogues with Yields and Purification Methods Highlighted for Key Intermediates
Figure 2
(a) SAM and SeAM analogues examined as cofactor surrogates for native and engineered PMTs. (b) Relative transalkylation efficiency (S vs Se) on known peptide substrates for selected engineered PMTs and SAM/SeAM analogues. Degrees of alkylation (equivalent to units of the consumed cofactor) per unit peptide were quantified via MALDI-MS (see the Supporting Information for more details). The difference between SeAM and their equivalent SAM analogues were then compared and categorized. NR, no reaction; =, no observable difference; +, an increase of 0.1–0.5 equiv alkylation/substrate; ++, an increase of >0.5 equiv alkylation/substrate; −, a decrease of 0.1–0.5 equiv alkylation/substrate from _S_- to _S_e-alkyl SAM analogues. Relative values for units of the consumed cofactors per unit peptide substrate are shown in parentheses as (_S_-analogue, _Se_-analogue) (c, d) Representative MALDI-MS analysis: PRMT3M233G-catalyzed reactions using hexyne–SAM and −SeAM cofactors (13a vs 13b); G9a Y1154A-catalyzed reactions using enyne–SAM and −SeAM cofactors (12a vs 12b).
Similar articles
- Se-adenosyl-L-selenomethionine cofactor analogue as a reporter of protein methylation.
Bothwell IR, Islam K, Chen Y, Zheng W, Blum G, Deng H, Luo M. Bothwell IR, et al. J Am Chem Soc. 2012 Sep 12;134(36):14905-12. doi: 10.1021/ja304782r. Epub 2012 Sep 4. J Am Chem Soc. 2012. PMID: 22917021 Free PMC article. - An efficient method for the synthesis of selenium modified nucleosides: its application in the synthesis of Se-adenosyl-L-selenomethionine (SeAM).
Kogami M, Koketsu M. Kogami M, et al. Org Biomol Chem. 2015 Sep 28;13(36):9405-17. doi: 10.1039/c5ob01316j. Epub 2015 Aug 6. Org Biomol Chem. 2015. PMID: 26246151 - Propargylic _Se_-adenosyl-l-selenomethionine: A Chemical Tool for Methylome Analysis.
Sohtome Y, Shimazu T, Shinkai Y, Sodeoka M. Sohtome Y, et al. Acc Chem Res. 2021 Oct 19;54(20):3818-3827. doi: 10.1021/acs.accounts.1c00395. Epub 2021 Oct 6. Acc Chem Res. 2021. PMID: 34612032 Review. - Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine.
Iwig DF, Booker SJ. Iwig DF, et al. Biochemistry. 2004 Oct 26;43(42):13496-509. doi: 10.1021/bi048693+. Biochemistry. 2004. PMID: 15491157 - Synthesis of selenocysteine and selenomethionine derivatives from sulfur-containing amino acids.
Iwaoka M, Ooka R, Nakazato T, Yoshida S, Oishi S. Iwaoka M, et al. Chem Biodivers. 2008 Mar;5(3):359-74. doi: 10.1002/cbdv.200890037. Chem Biodivers. 2008. PMID: 18357559 Review.
Cited by
- Analogs of _S_-Adenosyl-_L_-Methionine in Studies of Methyltransferases.
Rudenko AY, Mariasina SS, Sergiev PV, Polshakov VI. Rudenko AY, et al. Mol Biol. 2022;56(2):229-250. doi: 10.1134/S002689332202011X. Epub 2022 Apr 14. Mol Biol. 2022. PMID: 35440827 Free PMC article. - Profiling and Validation of Live-Cell Protein Methylation with Engineered Enzymes and Methionine Analogues.
Weiss N, Seneviranthe C, Jiang M, Wang K, Luo M. Weiss N, et al. Curr Protoc. 2021 Aug;1(8):e213. doi: 10.1002/cpz1.213. Curr Protoc. 2021. PMID: 34370893 Free PMC article. - A metabolic labeling method detects m6A transcriptome-wide at single base resolution.
Shu X, Cao J, Cheng M, Xiang S, Gao M, Li T, Ying X, Wang F, Yue Y, Lu Z, Dai Q, Cui X, Ma L, Wang Y, He C, Feng X, Liu J. Shu X, et al. Nat Chem Biol. 2020 Aug;16(8):887-895. doi: 10.1038/s41589-020-0526-9. Epub 2020 Apr 27. Nat Chem Biol. 2020. PMID: 32341503 - Synthesis and characterization of _Se_-adenosyl-L-selenohomocysteine selenoxide.
Duclos RI Jr, Cleary DC, Catcott KC, Zhou ZS. Duclos RI Jr, et al. J Sulphur Chem. 2015 Apr 1;36(2):135-144. doi: 10.1080/17415993.2014.979173. J Sulphur Chem. 2015. PMID: 26005494 Free PMC article. - Synthesis and biological evaluation of novel curcuminoid derivatives.
Cao YK, Li HJ, Song ZF, Li Y, Huai QY. Cao YK, et al. Molecules. 2014 Oct 13;19(10):16349-72. doi: 10.3390/molecules191016349. Molecules. 2014. PMID: 25314599 Free PMC article.
References
- Cantoni G. L. J. Biol. Chem. 1953, 204, 403. - PubMed
- Zhang L.; Ding X. J.; Cui J.; Xu H.; Chen J.; Gong Y. N.; Hu L. Y.; Zhou Y.; Ge J. N.; Lu Q. H.; Liu L. P.; Chen S.; Shao F. Nature 2012, 481, 204. - PubMed
- Van Der Werf P.; Koshland D. E. Jr. J. Biol. Chem. 1977, 252, 2793. - PubMed
- Rea S.; Eisenhaber F.; O’Carroll N.; Strahl B. D.; Sun Z. W.; Schmid M.; Opravil S.; Mechtler K.; Ponting C. P.; Allis C. D.; Jenuwein T. Nature 2000, 406, 593. - PubMed
- Bernstein B. E.; Meissner A.; Lander E. S. Cell 2007, 128, 669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources