In silico docking, molecular dynamics and binding energy insights into the bolinaquinone-clathrin terminal domain binding site - PubMed (original) (raw)
In silico docking, molecular dynamics and binding energy insights into the bolinaquinone-clathrin terminal domain binding site
Mohammed K Abdel-Hamid et al. Molecules. 2014.
Abstract
Clathrin-mediated endocytosis (CME) is a process that regulates selective internalization of important cellular cargo using clathrin-coated vesicles. Perturbation of this process has been linked to many diseases including cancer and neurodegenerative conditions. Chemical proteomics identified the marine metabolite, 2-hydroxy-5-methoxy-3-(((1S,4aS,8aS)-1,4a,5-trimethyl-1,2,3,4,4a,7,8,8a-octahydronaphthalen-2-yl)methyl)cyclohexa- 2,5-diene-1,4-dione (bolinaquinone) as a clathrin inhibitor. While being an attractive medicinal chemistry target, the lack of data about bolinaquinone's mode of binding to the clathrin enzyme represents a major limitation for its structural optimization. We have used a molecular modeling approach to rationalize the observed activity of bolinaquinone and to predict its mode of binding with the clathrin terminal domain (CTD). The applied protocol started by global rigid-protein docking followed by flexible docking, molecular dynamics and linear interaction energy calculations. The results revealed the potential of bolinaquinone to interact with various pockets within the CTD, including the clathrin-box binding site. The results also highlight the importance of electrostatic contacts over van der Waals interactions for proper binding between bolinaquinone and its possible binding sites. This study provides a novel model that has the potential to allow rapid elaboration of bolinaquinone analogues as a new class of clathrin inhibitors.
Conflict of interest statement
The authors declare the following competing financial interest(s): We have entered into a commercial agreement with Abcam Biochemicals (Bristol, UK) for the supply of our clathrin inhibitors. This includes some of the compounds listed in this paper. Pitstop® is a registered trademark of Children’s Medical Research Institute (Sydney, Australia) and Newcastle Innovation, Ltd. (Newcastle, Australia)
Figures
Figure 1
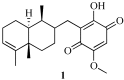
Chemical structure of the marine metabolite bolinaquinone (1).
Figure 2
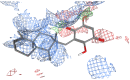
(A) Ribbon representation of a top view of the CTD (2XZG). The positioning of each of the seven β-stranded blades is indicated by the numbers 1-7. The predicted binding clusters for the docked 1 poses are rendered in stick representation and are color coded (non-carbon atoms) with  (clathrin-box binding site),
(clathrin-box binding site),  (site 1),
(site 1),  (site 2) and
(site 2) and  (site 3). (B) Same representation as in A rotated 90° inward, indicating potential 1 binding sites. (C) Bar graph representation of the number of 1 poses in each binding site cluster shown in A and B. The bar colors correspond to the binding sites and 1-clusters identified in A and B.
(site 3). (B) Same representation as in A rotated 90° inward, indicating potential 1 binding sites. (C) Bar graph representation of the number of 1 poses in each binding site cluster shown in A and B. The bar colors correspond to the binding sites and 1-clusters identified in A and B.
Figure 3
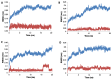
Molecular dynamics trajectory plots correlating RMSD deviation from the initial 1-CTD protein Cα atoms (blue) and 1 heavy atoms (red) coordinates over a simulation time of 20 ns. (A) Trajectory plot output for binding site 1; (B) Trajectory plot output for binding site 2; (C) Trajectory plot output for binding site 3; and (D) Trajectory plot output for the clathrin-box binding site. Stable complexes are shown in A, B and D while C shows distortion of the complex structure just after 15 ns.
Figure 4
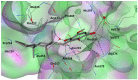
The average energy minimized structure of the molecular dynamics simulated 1 (grey stick representation) bound at the potential binding site 1 of the CTD (only relevant residues are shown as line representation and water molecules are shown as red spheres). Potential hydrogen bonding between 1 and the binding site are shown as black dashed lines.
Figure 5
Electrostatics maps (Hydrophobic; green, Hydrogen bond acceptor; red and Hydrogen bond donor; blue) for 1 (stick representation) bound at the proposed CTD binding site.
Similar articles
- Chemical proteomics reveals bolinaquinone as a clathrin-mediated endocytosis inhibitor.
Margarucci L, Monti MC, Fontanella B, Riccio R, Casapullo A. Margarucci L, et al. Mol Biosyst. 2011 Feb;7(2):480-5. doi: 10.1039/c0mb00126k. Epub 2010 Nov 8. Mol Biosyst. 2011. PMID: 21060934 - Identifying Small-Molecule Inhibitors of the Clathrin Terminal Domain.
Haucke V, Krauß M. Haucke V, et al. Methods Mol Biol. 2018;1847:51-64. doi: 10.1007/978-1-4939-8719-1_5. Methods Mol Biol. 2018. PMID: 30129009 - Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs.
Collette JR, Chi RJ, Boettner DR, Fernandez-Golbano IM, Plemel R, Merz AJ, Geli MI, Traub LM, Lemmon SK. Collette JR, et al. Mol Biol Cell. 2009 Jul;20(14):3401-13. doi: 10.1091/mbc.e08-10-1082. Epub 2009 May 20. Mol Biol Cell. 2009. PMID: 19458198 Free PMC article. - Clathrin-protein interactions.
Lafer EM. Lafer EM. Traffic. 2002 Aug;3(8):513-20. doi: 10.1034/j.1600-0854.2002.30801.x. Traffic. 2002. PMID: 12121414 Review. - Linking endocytic cargo to clathrin: structural and functional insights into coated vesicle formation.
Owen DJ. Owen DJ. Biochem Soc Trans. 2004 Feb;32(Pt 1):1-14. doi: 10.1042/bst0320001. Biochem Soc Trans. 2004. PMID: 14748702 Review.
Cited by
- Multiscale Simulations of Biological Membranes: The Challenge To Understand Biological Phenomena in a Living Substance.
Enkavi G, Javanainen M, Kulig W, Róg T, Vattulainen I. Enkavi G, et al. Chem Rev. 2019 May 8;119(9):5607-5774. doi: 10.1021/acs.chemrev.8b00538. Epub 2019 Mar 12. Chem Rev. 2019. PMID: 30859819 Free PMC article. - Pharmacological evaluation of newly synthesized organotin IV complex for antiulcer potential.
Azmatullah S, Khan AU, Qazi NG, Nadeem H, Irshad N. Azmatullah S, et al. BMC Pharmacol Toxicol. 2022 Jul 29;23(1):58. doi: 10.1186/s40360-022-00596-0. BMC Pharmacol Toxicol. 2022. PMID: 35906691 Free PMC article. - The Chemical Inhibitors of Endocytosis: From Mechanisms to Potential Clinical Applications.
Szewczyk-Roszczenko OK, Roszczenko P, Shmakova A, Finiuk N, Holota S, Lesyk R, Bielawska A, Vassetzky Y, Bielawski K. Szewczyk-Roszczenko OK, et al. Cells. 2023 Sep 19;12(18):2312. doi: 10.3390/cells12182312. Cells. 2023. PMID: 37759535 Free PMC article. Review. - The sulfonadyns: a class of aryl sulfonamides inhibiting dynamin I GTPase and clathrin mediated endocytosis are anti-seizure in animal models.
Odell LR, Jones NC, Chau N, Robertson MJ, Ambrus JI, Deane FM, Young KA, Whiting A, Xue J, Prichard K, Daniel JA, Gorgani NN, O'Brien TJ, Robinson PJ, McCluskey A. Odell LR, et al. RSC Med Chem. 2023 Apr 26;14(8):1492-1511. doi: 10.1039/d2md00371f. eCollection 2023 Aug 16. RSC Med Chem. 2023. PMID: 37593570 Free PMC article. - Ebastine impairs metastatic spread in triple-negative breast cancer by targeting focal adhesion kinase.
Seo J, Park M, Ko D, Kim S, Park JM, Park S, Nam KD, Farrand L, Yang J, Seok C, Jung E, Kim YJ, Kim JY, Seo JH. Seo J, et al. Cell Mol Life Sci. 2023 Apr 25;80(5):132. doi: 10.1007/s00018-023-04760-5. Cell Mol Life Sci. 2023. PMID: 37185776 Free PMC article.
References
- Von Kleist L., Stahlschmidt W., Bulut H., Gromova K., Puchkov D., Robertson M.J., MacGregor K.A., Tomilin N., Pechstein A., Chau N., et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146:471–484. doi: 10.1016/j.cell.2011.06.025. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources