Targeted gene therapy and cell reprogramming in Fanconi anemia - PubMed (original) (raw)
doi: 10.15252/emmm.201303374. Epub 2014 Apr 6.
Rocio Baños 1, Angelo Lombardo 2, Oscar Quintana-Bustamante 1, Lara Alvarez 1, Zita Garate 1, Pietro Genovese 2, Elena Almarza 1, Antonio Valeri 1, Begoña Díez 1, Susana Navarro 1, Yaima Torres 3, Juan P Trujillo 4, Rodolfo Murillas 5, Jose C Segovia 1, Enrique Samper 3, Jordi Surralles 6, Philip D Gregory 7, Michael C Holmes 7, Luigi Naldini 8, Juan A Bueren 9
Affiliations
- PMID: 24859981
- PMCID: PMC4203359
- DOI: 10.15252/emmm.201303374
Targeted gene therapy and cell reprogramming in Fanconi anemia
Paula Rio et al. EMBO Mol Med. 2014 Jun.
Abstract
Gene targeting is progressively becoming a realistic therapeutic alternative in clinics. It is unknown, however, whether this technology will be suitable for the treatment of DNA repair deficiency syndromes such as Fanconi anemia (FA), with defects in homology-directed DNA repair. In this study, we used zinc finger nucleases and integrase-defective lentiviral vectors to demonstrate for the first time that FANCA can be efficiently and specifically targeted into the AAVS1 safe harbor locus in fibroblasts from FA-A patients. Strikingly, up to 40% of FA fibroblasts showed gene targeting 42 days after gene editing. Given the low number of hematopoietic precursors in the bone marrow of FA patients, gene-edited FA fibroblasts were then reprogrammed and re-differentiated toward the hematopoietic lineage. Analyses of gene-edited FA-iPSCs confirmed the specific integration of FANCA in the AAVS1 locus in all tested clones. Moreover, the hematopoietic differentiation of these iPSCs efficiently generated disease-free hematopoietic progenitors. Taken together, our results demonstrate for the first time the feasibility of correcting the phenotype of a DNA repair deficiency syndrome using gene-targeting and cell reprogramming strategies.
Keywords: Fanconi anemia; cell reprogramming; gene‐targeting; iPSCs; zinc finger nucleases.
© 2014 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
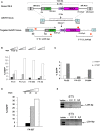
Figure 1. Efficacy of gene targeting of FANCA in the AAVS1 locus of primary hFA-A fibroblasts
- Top: schematic representation of the donor integrase-defective lentiviral vector (IDLV) used to promote insertion of the EGFP/FANCA cassette into the AAVS1 locus. Middle: AAVS1 locus with the zinc finger nucleases (ZFNs) target site. Bottom: AAVS1 locus upon ZFN-mediated targeted insertion of the EGFP/PGK-FANCA cassette. Black arrow shows transcription of the EGFP from the endogenous PPP1R12C promoter. HA, homology arm; SD, splice donor; SA, splice acceptor; BGHpA, bovine growth hormone polyadenylation signal; SV40pA, simian virus 40 polyadenylation signal. Constituents of the LTR (U5-R-ΔU3) are also indicated.
- Proliferation advantage of targeted Fanconi anemia (FA) fibroblasts (EGFP+ cells) during in vitro incubation.
- Comparative analysis of gene targeting in FA-A fibroblasts, untransduced or transduced with a lentiviral vector expressing h_TERT_. Analyses were performed 14 days after gene targeting.
- In vitro proliferation advantage of targeted FA fibroblasts (EGFP+) previously transduced with hTERT (FA-52T fibroblasts).
- Targeted integration analysis of the EGFP/PGK-FANCA cassette into the AAVS1 site by PCR using primers specific for the 5′ or 3′ integration junctions (red arrows in the top schematic) defined as 5′ TI or 3′ TI, respectively.
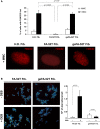
Figure 2. Phenotypic correction of the gene-edited FA-A fibroblasts
- Top: histogram showing the percentage of FA-A fibroblasts, unstransduced or co-transduced with the donor integrase-defective lentiviral vector (IDLV) and the AdV5/35-ZFNs (geFA-52T Fib), showing FANCD2 foci in the absence or the presence of mitomycin C (MMC). Bottom: representative images of FANCD2 foci (red) in cells shown in the top histogram, after MMC treatment.
- Chromosomal instability induced by diepoxybutane (DEB) in untreated (FA-52T) and gene-edited FA fibroblasts (geFA-52T Fib). Left: representative FISH analysis was performed by staining telomeres (in green), centromeres (in pink) and chromosomes (in blue). Right: histogram showing the number of chromosomal aberrations per cell.
Data information: Values are shown as mean ± s.e. from three independent experiments (A) or analysis of twenty different metaphases per group (B). All _P_-values were calculated using two-tailed unpaired Student's _t_-test.
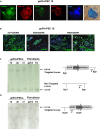
Figure 3. Pluripotency characterization and insertion site analyses of gene-edited FA-A iPSCs
- Expression of TRA1-60, SSEA-4, OCT4, and NANOG pluripotency markers by immunofluorescence staining of gene-edited FA-iPSCs (geFA-iPSCs; clone 16).
- Immunofluorescence analysis of ectoderm (β-II-tubulin), endoderm (Fox2A), and mesoderm (α-SMA and Brachyury) in teratomas generated from geFA-iPSCs (clone 16).
- Southern blot analysis of genomic DNA extracted from the indicated gene-corrected FA iPSC clones (geFA-IPSCs) and from parental fibroblasts, either unmanipulated (FA) or after gene editing (ge-FA iPSCs, clones 16, 26 and 31). Genomic DNA was digested with BglI and hybridized with a probe for PPP1R12C. The band of 9.6 kb corresponds to the targeted integration in PPP1R12C, while the 3.3 kb correspond to the untargeted allele.
- Southern blot analysis of samples shown in (C) digested with BstXI and hybridized with a probe (P) for EGFP. One single band of 5.1 kb is expected for specific integrations in PPP1R12C.
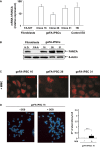
Figure 4. Disease-free Fanconi anemia phenotype of corrected geFA-iPSCs
- Histogram showing the levels of hFANCA expression in gene-edited FA-iPSC clones and human ES (H9) relative to untreated FA-52T fibroblasts. Data are shown as mean ± s.e. of three different analyses.
- Western blot analysis showing FANCA expression in geFA-iPSC clones in comparison with fibroblasts from HD and a FA-A patient.
- Representative immunofluorescence analysis of FANCD2 foci in geFA-iPSCs after DNA damage with mitomycin C (MMC).
- Chromosomal instability induced by diepoxybutane (DEB) was also tested in geFA-iPSC 16. FISH analysis was performed using probes to detect telomeres (green), centromeres (pink) and chromosomes (blue). Right: histogram showing the number of chromosomal aberrations per cell.
Data information: Data are shown as mean ± s.e. from three different experiments (A) or analysis of twenty different metaphases per group (D). All _P_-values were calculated using two-tailed unpaired Student's _t_-test.
Figure 5. Hematopoietic differentiation of gene-edited FA-IPSCs
- Analysis of the percentage of CD43+CD34+, CD45+CD34+, and CD45+ cells generated by unexcised and excised geFA-iPSCs (clones 16 and Ex 16.1).
- Left: Representative pictures of hematopoietic colonies generated by geFA-iPSCs. Right: Analysis of the clonogenic potential of unexcised and excised ge-FAiPSCs (clones 16 and Ex 16.1) in comparison with H.D. cord blood cells.
- Survival to mitomycin C (MMC) of CFCs obtained from geFA-IPSCs (clone 16) in comparison with BM CFCs from two different FA patients (FA-664 BM and FA-82 BM) and with CFCs from a healthy cord blood (H.D. CB).
Data information: Values are shown as mean ± s.e. of three experiments. All _P_-values were calculated using two-tailed unpaired Student's _t_-test.
Similar articles
- Overcoming reprogramming resistance of Fanconi anemia cells.
Müller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, Moreau LA, Schambach A, Park IH, London WB, Strait K, Schlaeger T, Devine AL, Grassman E, D'Andrea A, Daley GQ, Williams DA. Müller LU, et al. Blood. 2012 Jun 7;119(23):5449-57. doi: 10.1182/blood-2012-02-408674. Epub 2012 Feb 27. Blood. 2012. PMID: 22371882 Free PMC article. - Pluripotent cell models of fanconi anemia identify the early pathological defect in human hemoangiogenic progenitors.
Suzuki NM, Niwa A, Yabe M, Hira A, Okada C, Amano N, Watanabe A, Watanabe K, Heike T, Takata M, Nakahata T, Saito MK. Suzuki NM, et al. Stem Cells Transl Med. 2015 Apr;4(4):333-8. doi: 10.5966/sctm.2013-0172. Epub 2015 Mar 11. Stem Cells Transl Med. 2015. PMID: 25762002 Free PMC article. - Generation of iPSCs from genetically corrected Brca2 hypomorphic cells: implications in cell reprogramming and stem cell therapy.
Navarro S, Moleiro V, Molina-Estevez FJ, Lozano ML, Chinchon R, Almarza E, Quintana-Bustamante O, Mostoslavsky G, Maetzig T, Galla M, Heinz N, Schiedlmeier B, Torres Y, Modlich U, Samper E, Río P, Segovia JC, Raya A, Güenechea G, Izpisua-Belmonte JC, Bueren JA. Navarro S, et al. Stem Cells. 2014 Feb;32(2):436-46. doi: 10.1002/stem.1586. Stem Cells. 2014. PMID: 24420904 - Induced Pluripotency and Gene Editing in Fanconi Anemia.
Navarro S, Giorgetti A, Raya A, Tolar J. Navarro S, et al. Curr Gene Ther. 2017;16(5):321-328. doi: 10.2174/1566523217666170118112050. Curr Gene Ther. 2017. PMID: 28103772 Review. - Advances in Gene Therapy for Fanconi Anemia.
Río P, Navarro S, Bueren JA. Río P, et al. Hum Gene Ther. 2018 Oct;29(10):1114-1123. doi: 10.1089/hum.2018.124. Hum Gene Ther. 2018. PMID: 30117331 Review.
Cited by
- Clinically relevant gene editing in hematopoietic stem cells for the treatment of pyruvate kinase deficiency.
Fañanas-Baquero S, Quintana-Bustamante O, Dever DP, Alberquilla O, Sanchez-Dominguez R, Camarena J, Ojeda-Perez I, Dessy-Rodriguez M, Turk R, Schubert MS, Lattanzi A, Xu L, Lopez-Lorenzo JL, Bianchi P, Bueren JA, Behlke MA, Porteus M, Segovia JC. Fañanas-Baquero S, et al. Mol Ther Methods Clin Dev. 2021 May 14;22:237-248. doi: 10.1016/j.omtm.2021.05.001. eCollection 2021 Sep 10. Mol Ther Methods Clin Dev. 2021. PMID: 34485608 Free PMC article. - Therapy Development by Genome Editing of Hematopoietic Stem Cells.
Koniali L, Lederer CW, Kleanthous M. Koniali L, et al. Cells. 2021 Jun 14;10(6):1492. doi: 10.3390/cells10061492. Cells. 2021. PMID: 34198536 Free PMC article. Review. - Engineered Viruses as Genome Editing Devices.
Chen X, Gonçalves MA. Chen X, et al. Mol Ther. 2016 Mar;24(3):447-57. doi: 10.1038/mt.2015.164. Epub 2015 Sep 4. Mol Ther. 2016. PMID: 26336974 Free PMC article. Review. - CRISPR/Cas9 Targeted Gene Editing and Cellular Engineering in Fanconi Anemia.
Osborn M, Lonetree CL, Webber BR, Patel D, Dunmire S, McElroy AN, DeFeo AP, MacMillan ML, Wagner J, Balzar BR, Tolar J. Osborn M, et al. Stem Cells Dev. 2016 Oct;25(20):1591-1603. doi: 10.1089/scd.2016.0149. Epub 2016 Aug 18. Stem Cells Dev. 2016. PMID: 27538887 Free PMC article. - Mechanisms underlying the formation of induced pluripotent stem cells.
González F, Huangfu D. González F, et al. Wiley Interdiscip Rev Dev Biol. 2016 Jan-Feb;5(1):39-65. doi: 10.1002/wdev.206. Epub 2015 Sep 18. Wiley Interdiscip Rev Dev Biol. 2016. PMID: 26383234 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous