Lessons from miR-143/145: the importance of cell-type localization of miRNAs - PubMed (original) (raw)
Review
. 2014 Jul;42(12):7528-38.
doi: 10.1093/nar/gku461. Epub 2014 May 29.
Affiliations
- PMID: 24875473
- PMCID: PMC4081080
- DOI: 10.1093/nar/gku461
Review
Lessons from miR-143/145: the importance of cell-type localization of miRNAs
Oliver A Kent et al. Nucleic Acids Res. 2014 Jul.
Abstract
miR-143 and miR-145 are co-expressed microRNAs (miRNAs) that have been extensively studied as potential tumor suppressors. These miRNAs are highly expressed in the colon and are consistently reported as being downregulated in colorectal and other cancers. Through regulation of multiple targets, they elicit potent effects on cancer cell growth and tumorigenesis. Importantly, a recent discovery demonstrates that miR-143 and miR-145 are not expressed in colonic epithelial cells; rather, these two miRNAs are highly expressed in mesenchymal cells such as fibroblasts and smooth muscle cells. The expression patterns of miR-143 and miR-145 and other miRNAs were initially determined from tissue level data without consideration that multiple different cell types, each with their own unique miRNA expression patterns, make up each tissue. Herein, we discuss the early reports on the identification of dysregulated miR-143 and miR-145 expression in colorectal cancer and how lack of consideration of cellular composition of normal tissue led to the misconception that these miRNAs are downregulated in cancer. We evaluate mechanistic data from miR-143/145 studies in context of their cell type-restricted expression pattern and the potential of these miRNAs to be considered tumor suppressors. Further, we examine other examples of miRNAs being investigated in inappropriate cell types modulating pathways in a non-biological fashion. Our review highlights the importance of determining the cellular expression pattern of each miRNA, so that downstream studies are conducted in the appropriate cell type.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
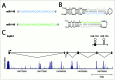
Figure 1.
The miR-143/145 cluster. (A) Mature miRNA sequences of miR-143 and miR-145. (B) The proposed secondary structures of the pre-miR-143 and pre-miR-145 stem loops. The mature miRNA sequences are blue (miR-143) and green (miR-145). miRNA-star sequences are shown in gray. (C) The genomic organization and major primary transcript structure of the miR-143/145 cluster. The minor transcript (not shown) lacks exon 2. The plot depicted below the transcript shows evolutionary conservation (UCSC Genome Browser 28 species conservation track, NCBI36/hg18 assembly).
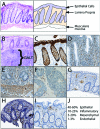
Figure 2.
Cellular components of the colonic mucosa with direct visualization of unique cell types by immunohistochemistry. (A) Representative hematoxylin & eosin image of a typical normal colonic biopsy (left) and a schematic representation of the colon biopsy (right) indicating cell types found in the tissue. (B) Normal gut-associated lymphoid tissue (GALT) containing collections of lymphocytes can be found in the mucosa and superficial submucosa of some biopsies. (C) AE1/AE3 staining of the epithelial component of the colonic mucosa. (D) Smooth muscle actin (SMA) staining of normal colon demonstrating SMC, pericyte and fibroblast staining within the lamina propria and (E) more extensive SMA staining of SMCs in the muscularis mucosa. (F) SMA staining in an exophytic colon adenocarcinoma is greatly reduced compared to normal tissue. (G) CD34 staining demonstrating endothelial cells within the lamina propria. (H) CD3 staining demonstrating T lymphocytes scattered within the lamina propria. (I) CD68 staining demonstrating predominantly macrophages underlying the surface epithelium. (J) Image analysis of these staining patterns across multiple routine colonic biopsies identified typical ratios of cellular composition of colon. These ratios can be markedly different depending on the amount of muscularis mucosa and GALT present in the biopsy.
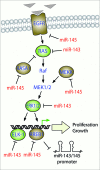
Figure 3.
miR-143/145 modulate signaling through the Ras-MAPK pathway. Extracellular mitogenic signals (triangles) leads to activation of Ras and the signaling through the Raf-MEK-ERK (MAPK) cascade to activate transcription factors such as Elk-1 and RREB1 resulting in proliferation and growth responses. Targets of miR-143 and miR-145 are shown in green and brown respectively. The miR-143/145 proximal promoter is negatively regulated by K-Ras-RREB1 feedback loop.
Similar articles
- Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Gunaratne PH. Gunaratne PH. Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review. - Entamoeba histolytica Up-Regulates MicroRNA-643 to Promote Apoptosis by Targeting XIAP in Human Epithelial Colon Cells.
López-Rosas I, López-Camarillo C, Salinas-Vera YM, Hernández-de la Cruz ON, Palma-Flores C, Chávez-Munguía B, Resendis-Antonio O, Guillen N, Pérez-Plasencia C, Álvarez-Sánchez ME, Ramírez-Moreno E, Marchat LA. López-Rosas I, et al. Front Cell Infect Microbiol. 2019 Jan 8;8:437. doi: 10.3389/fcimb.2018.00437. eCollection 2018. Front Cell Infect Microbiol. 2019. PMID: 30671387 Free PMC article. - Small RNA expression from viruses, bacteria and human miRNAs in colon cancer tissue and its association with microsatellite instability and tumor location.
Mjelle R, Sjursen W, Thommesen L, Sætrom P, Hofsli E. Mjelle R, et al. BMC Cancer. 2019 Feb 20;19(1):161. doi: 10.1186/s12885-019-5330-0. BMC Cancer. 2019. PMID: 30786859 Free PMC article. - Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer.
Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, Bittner A, Raponi M. Arndt GM, et al. BMC Cancer. 2009 Oct 20;9:374. doi: 10.1186/1471-2407-9-374. BMC Cancer. 2009. PMID: 19843336 Free PMC article. - Modulatory roles of microRNAs in the regulation of different signalling pathways in large bowel cancer stem cells.
Mamoori A, Gopalan V, Smith RA, Lam AK. Mamoori A, et al. Biol Cell. 2016 Mar;108(3):51-64. doi: 10.1111/boc.201500062. Epub 2016 Feb 2. Biol Cell. 2016. PMID: 26712035 Review.
Cited by
- Toward the promise of microRNAs - Enhancing reproducibility and rigor in microRNA research.
Witwer KW, Halushka MK. Witwer KW, et al. RNA Biol. 2016 Nov;13(11):1103-1116. doi: 10.1080/15476286.2016.1236172. Epub 2016 Sep 19. RNA Biol. 2016. PMID: 27645402 Free PMC article. Review. - miRNA profiling of circulating EpCAM(+) extracellular vesicles: promising biomarkers of colorectal cancer.
Ostenfeld MS, Jensen SG, Jeppesen DK, Christensen LL, Thorsen SB, Stenvang J, Hvam ML, Thomsen A, Mouritzen P, Rasmussen MH, Nielsen HJ, Ørntoft TF, Andersen CL. Ostenfeld MS, et al. J Extracell Vesicles. 2016 Aug 29;5:31488. doi: 10.3402/jev.v5.31488. eCollection 2016. J Extracell Vesicles. 2016. PMID: 27576678 Free PMC article. - Complex Sources of Variation in Tissue Expression Data: Analysis of the GTEx Lung Transcriptome.
McCall MN, Illei PB, Halushka MK. McCall MN, et al. Am J Hum Genet. 2016 Sep 1;99(3):624-635. doi: 10.1016/j.ajhg.2016.07.007. Am J Hum Genet. 2016. PMID: 27588449 Free PMC article. - miR-143 Regulates Lysosomal Enzyme Transport across the Blood-Brain Barrier and Transforms CNS Treatment for Mucopolysaccharidosis Type I.
Lin Y, Wang X, Rose KP, Dai M, Han J, Xin M, Pan D. Lin Y, et al. Mol Ther. 2020 Oct 7;28(10):2161-2176. doi: 10.1016/j.ymthe.2020.06.011. Epub 2020 Jun 15. Mol Ther. 2020. PMID: 32610100 Free PMC article. - Let-7f: A New Potential Circulating Biomarker Identified by miRNA Profiling of Cells Isolated from Human Abdominal Aortic Aneurysm.
Spear R, Boytard L, Blervaque R, Chwastyniak M, Hot D, Vanhoutte J, Lamblin N, Amouyel P, Pinet F. Spear R, et al. Int J Mol Sci. 2019 Nov 5;20(21):5499. doi: 10.3390/ijms20215499. Int J Mol Sci. 2019. PMID: 31694153 Free PMC article.
References
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources