Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization - PubMed (original) (raw)
doi: 10.1038/nm.3563. Epub 2014 Jun 1.
Nandi Simpson 2, Olivier Join-Lambert 3, Christian Federici 4, Marie-Pierre Laran-Chich 4, Nawal Maïssa 4, Haniaa Bouzinba-Ségard 4, Philippe C Morand 5, Fabrice Chretien 6, Saïd Taouji 7, Eric Chevet 7, Sébastien Janel 8, Frank Lafont 8, Mathieu Coureuil 9, Audrey Segura 9, Florence Niedergang 4, Stefano Marullo 4, Pierre-Olivier Couraud 4, Xavier Nassif 3, Sandrine Bourdoulous 4
Affiliations
- PMID: 24880614
- PMCID: PMC7095922
- DOI: 10.1038/nm.3563
Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization
Sandra C Bernard et al. Nat Med. 2014 Jul.
Abstract
Neisseria meningitidis is a cause of meningitis epidemics worldwide and of rapidly progressing fatal septic shock. A crucial step in the pathogenesis of invasive meningococcal infections is the adhesion of bloodborne meningococci to both peripheral and brain endothelia, leading to major vascular dysfunction. Initial adhesion of pathogenic strains to endothelial cells relies on meningococcal type IV pili, but the endothelial receptor for bacterial adhesion remains unknown. Here, we report that the immunoglobulin superfamily member CD147 (also called extracellular matrix metalloproteinase inducer (EMMPRIN) or Basigin) is a critical host receptor for the meningococcal pilus components PilE and PilV. Interfering with this interaction potently inhibited the primary attachment of meningococci to human endothelial cells in vitro and prevented colonization of vessels in human brain tissue explants ex vivo and in humanized mice in vivo. These findings establish the molecular events by which meningococci target human endothelia, and they open new perspectives for treatment and prevention of meningococcus-induced vascular dysfunctions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
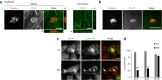
Figure 1. CD147 accumulates at sites of meningococcal adhesion independently of β2-AR activation.
(a) CD147 staining in noninfected and infected polarized monolayer of hCMEC/D3 cells. CD147 (green) and meningococcal colonies (red) in reconstructed images of 2-h infection; image reconstructions along xz and yz axes are shown. Scale bars, 10 μm. (b) Magnification of an adherent diplococcus 10 min after infection. Scale bar, 5 μm. (c) hCMEC/D3 cells expressing exogenous YFP-tagged β2-AR (β2-AR–YFP) left untreated (NT) or preincubated for 1 h with 10 μM isoproterenol (ISO) to induce endocytosis of the β2-AR before 2-h infection with wild-type meningococci. Arrows indicate the location of adherent meningococcus colonies. Scale bars, 10 μm. Images are representative of 3 independent experiments. (d) Percentage of the bacterial colonies associated with ezrin, CD44 or CD147 recruitment to sites of meningococcal adhesion in untreated cells or after cell pretreatment with 10 μM isoproterenol to promote β2-AR endocytosis. Mean ± s.e.m.; n = 3 experiments; ***P < 0.001, two-way analysis of variance (ANOVA).
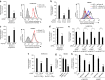
Figure 2. CD147 depletion, soluble CD147 and anti-CD147 antibodies inhibit N. meningitidis adhesion to human endothelial cells.
(a) Left, adhesion of Nm2C4.3 to CD147-depleted (siRNA CD147) or control (siRNA CTL) hCMEC/D3 cells, quantified following a 30-min infection. Right, FACS analysis of CD147 expression in hCMEC/D3 cells, with isotype control as shaded area and mean fluorescence intensities in parentheses. Mean ± s.e.m., n = 4; **P < 0.05, two-tailed Student's _t_-test. (b) Left, adhesion of Nm2C4.3 to control hBMEC cells (siRNA CTL + empty vector), CD147-depleted (siRNA CD147 + empty vector) or CD147-complemented HBMECs (siRNA CD147 + plasmid encoding CD147), quantified following a 30-min infection. Right, FACS analysis of CD147 expression in HBMEC cells, with isotype control as shaded area and mean fluorescence intensities in parentheses. **P = 0.0025, two-tailed Student's _t_-test. (c) Left, adhesion of Nm2C4.3 to control (empty plasmid) or CD147-overexpressing (CD147 plasmid) hBMEC cells, quantified following a 30-min infection. Right, FACS analysis of CD147 expression in HBMEC cells, with isotype control as shaded area and mean fluorescence intensities in parentheses. Mean ± s.e.m., n = 3; **P = 0.0094, two-tailed Student's _t_-test. (d) Adhesion of Nm2C4.3 to CD147-depleted or control hCMEC/D3 cells, quantified following a 10-min infection under shear stress (0.04 dynes/cm2). **P < 0.01, two-tailed Student's _t_-test. (e) Adhesion of meningococcus strains belonging to different sequence types and capsular serotypes to CD147-depleted or control hCMEC/D3 cells, quantified following a 30-min infection. Mean ± s.e.m., n = 4; *P = 0.025, **P = 0.001, ***P = 0.0008, two-tailed Student's _t_-test. (f) Adhesion of Nm2C4.3 pilin variant strains expressing PilESA or PilESB to CD147-depleted or control hCMEC/D3 cells, quantified following a 30-min infection. Mean ± s.e.m., n = 6; ***P < 0.001, two-way ANOVA. (g) Adhesion of Nm2C4.3 to HBMECs in the absence (negative) or in the presence of purified CD147-Fc or ICAM-1–Fc (both at 5 μg ml−1), quantified following a 30-min infection. **P < 0.01, one-way ANOVA. (h) Adhesion of Nm2C4.3 to HBMECs and hCMEC/D3 cells preincubated with 10 μg ml−1 of antibodies targeting ICAM-1 (11C81), the N-terminal Ig domain of CD147 (MEM-M6/1) or the C-terminal Ig domain of CD147 (MEM-M6/6), quantified following a 10-min infection under shear stress (0.04 dynes/cm2). Abs, antibodies. Mean ± s.e.m., n = 3; **P < 0.01, ***P < 0.001, one-way ANOVA.
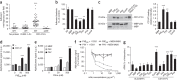
Figure 3. The major pilin PilE and the minor pilin PilV interact directly with CD147.
(a) Adhesion of wild-type (WT) and nonpiliated (Δ_pilE_, as control) meningococci to immobilized CD147-Fc, or to ICAM-1–Fc or no immobilized protein as controls, quantified following a 1-h infection. Mean ± s.e.m., n = 4; ***P < 0.001, one-way ANOVA. (b) Adhesion of Nm2C4.3 to hCMEC/D3 cells in the absence (Nontreated, NT) or in the presence of soluble MBP-pilins (PilESA, PilESB, PilV, PilX or ComP) or MBP as a control, quantified following a 30-min infection. Mean ± s.e.m., n = 4; **P < 0.01, ***P < 0.001, one-way ANOVA. (c) Left, CD147-Fc coprecipitation with staphylococci conjugated with the indicated MBP-pilins, or MBP alone as a control. Right, quantification of CD147 immunoprecipitation. Mean ± s.e.m., n = 4; **P < 0.01, ***P < 0.001, one-way ANOVA. (d) AlphaScreen analysis of the interaction between CD147-Fc (500 nM) or vehicle (−) as a control, and increasing MBP-PilESB concentrations. Mean ± s.e.m., n = 2; ***P < 0.001, two-way ANOVA. (e) AlphaScreen analysis of the interaction between CD147-Fc (500 nM) and MBP, MBP-PilX or MBP-PilV at various concentrations. Mean ± s.e.m., n = 2 experiments; ***P < 0.001, two-way ANOVA. (f) AlphaScreen analysis of the interaction between CD147-Fc (100 nM) and MBP-PilESB or MBP-PilV (500 nM) in the presence of increasing concentration of the anti-CD147 antibody (MEM-M6/6) or anti–ICAM-1 (11C81, as control). mAb, monoclonal antibody. (g) AFM analysis of the retraction forces between cantilever tips functionalized with MBP, MBP-pilins (PilX, PilV, PilESA and PilESB) or cyclophilin (Cyclo, positive control) and CD147-Fc or activated leukocyte-cell adhesion molecule-1 (ALCAM-1)-Fc (control). Bar plots indicate the percentage of force curves with interaction events. Mean ± s.e.m.; n = 7. ***P < 0.001, one-way ANOVA.
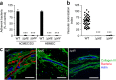
Figure 4. The major pilin PilE and the minor pilin PilV are essential bacterial components for vascular colonization.
(a) Adhesion of Nm2C4.3 wild-type strain or Δ_pilE_ and Δ_pilV_ mutant derivatives to hCMEC/D3 cells and HBMECs. Mean ± s.e.m., n = 4; ***P < 0.001, one-way ANOVA. (b,c) Colonization of human vessels in grafted human skin by Nm2C4.3 wild-type strain or the Δ_pilE_ or the Δ_pilV_ mutant derivatives. (b) Mean ± s.e.m. of the vascular colonization index. Analysis of 10 sections per graft, n = 6 grafts per condition. ***P < 0.001, one-way ANOVA. (c) Representative fluorescence microscopy showing the different bacterial derivatives (red) associated with human vessels (basal lamina collagen IV, green) in the skin. Scale bars, 100 μm. Images are representative of 6 different human skin grafts.
Figure 5. In situ interaction of N. meningitidis with human brain vessels requires both PilE and PilV expression.
(a) Immunohistochemical analysis of bacteria (brown) in a whole human brain section infected with Nm2C4.3 wild-type strain. Arrows indicate bacterial aggregates within brain vessels in the cortical region. Scale bar, 1 mm. (b) Detailed immunohistochemical analysis of the bacterial staining (blue) in the cortical region of serial human brain sections from the same donor infected with Nm2C4.3 wild-type strain or the Δ_pilE_ or the Δ_pilV_ mutant derivatives. Scale bar, 100 μm. (c) Detailed immunofluorescence analysis of bacteria (red), actin (green) and nucleus (blue) in the meningeal region (Virchow-Robin space) of serial human brain sections from the same donor infected with Nm2C4.3 wild-type strain or the Δ_pilE_ or the Δ_pilV_ mutant derivatives. Scale bars, 100 μm. (d) Quantification of adherent meningococci per 100-μm2 section, expressed as percentage of adherent wild-type bacteria. Mean ± s.e.m., analysis of 3 brain sections per brain from 3 different donors; ***P < 0.001, one-way ANOVA.
Figure 6. In situ infection of fresh human brain sections by N. meningitidis requires pilus interaction with CD147.
(a) Immunofluorescence analysis of CD147 (green), meningococcus colonies (red) and nucleus (blue) in the Virchow-Robin space and cortical region of serial brain sections from the same donor infected with Nm2C4.3 wild-type strain for 1 h. Scale bars, 20 μm. Images are representative of 6 different human skin grafts. (b) Immunohistochemical (top) and immunofluorescence (bottom) analysis of serial sections of cortical regions of the same donor, treated with 10 μg ml−1 anti–ICAM-1 (11C81) or anti-CD147 (MEM-M6/1 and MEM-M6/6) antibodies before infection with Nm2C4.3 wild-type meningococci. Scale bars, 100 μm (top) and 20 μm (bottom). Images are representative of 3 different human skin grafts. (c) Mean ± s.e.m. of total fluorescence per 100 μm2 relative to fluorescence in absence of antibody, n = 4 brain sections per donor from 3 different donors; *P < 0.05, one-way ANOVA.
Similar articles
- The minor pilin PilV provides a conserved adhesion site throughout the antigenically variable meningococcal type IV pilus.
Barnier JP, Meyer J, Kolappan S, Bouzinba-Ségard H, Gesbert G, Jamet A, Frapy E, Schönherr-Hellec S, Capel E, Virion Z, Dupuis M, Bille E, Morand P, Schmitt T, Bourdoulous S, Nassif X, Craig L, Coureuil M. Barnier JP, et al. Proc Natl Acad Sci U S A. 2021 Nov 9;118(45):e2109364118. doi: 10.1073/pnas.2109364118. Proc Natl Acad Sci U S A. 2021. PMID: 34725157 Free PMC article. - Type IV pilus retraction enables sustained bacteremia and plays a key role in the outcome of meningococcal sepsis in a humanized mouse model.
Barnier JP, Euphrasie D, Join-Lambert O, Audry M, Schonherr-Hellec S, Schmitt T, Bourdoulous S, Coureuil M, Nassif X, El Behi M. Barnier JP, et al. PLoS Pathog. 2021 Feb 16;17(2):e1009299. doi: 10.1371/journal.ppat.1009299. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33592056 Free PMC article. - Invasive meningococcal disease: a disease of the endothelial cells.
Coureuil M, Bourdoulous S, Marullo S, Nassif X. Coureuil M, et al. Trends Mol Med. 2014 Oct;20(10):571-8. doi: 10.1016/j.molmed.2014.08.002. Epub 2014 Aug 29. Trends Mol Med. 2014. PMID: 25178566 Review. - Strength of Neisseria meningitidis binding to endothelial cells requires highly-ordered CD147/β2-adrenoceptor clusters assembled by alpha-actinin-4.
Maïssa N, Covarelli V, Janel S, Durel B, Simpson N, Bernard SC, Pardo-Lopez L, Bouzinba-Ségard H, Faure C, Scott MGH, Coureuil M, Morand PC, Lafont F, Nassif X, Marullo S, Bourdoulous S. Maïssa N, et al. Nat Commun. 2017 Jun 1;8:15764. doi: 10.1038/ncomms15764. Nat Commun. 2017. PMID: 28569760 Free PMC article. - Meningococcal disease: A paradigm of type-IV pilus dependent pathogenesis.
Dos Santos Souza I, Maïssa N, Ziveri J, Morand PC, Coureuil M, Nassif X, Bourdoulous S. Dos Santos Souza I, et al. Cell Microbiol. 2020 Apr;22(4):e13185. doi: 10.1111/cmi.13185. Cell Microbiol. 2020. PMID: 32185901 Review.
Cited by
- Characterization of a novel antisense RNA in the major pilin locus of Neisseria meningitidis influencing antigenic variation.
Tan FY, Wörmann ME, Loh E, Tang CM, Exley RM. Tan FY, et al. J Bacteriol. 2015 May;197(10):1757-68. doi: 10.1128/JB.00082-15. Epub 2015 Mar 9. J Bacteriol. 2015. PMID: 25755192 Free PMC article. - Type IV pilus retraction is required for Neisseria musculi colonization and persistence in a natural mouse model of infection.
Rhodes KA, Rendón MA, Ma MC, Agellon A, Johnson AC, So M. Rhodes KA, et al. mBio. 2024 Jan 16;15(1):e0279223. doi: 10.1128/mbio.02792-23. Epub 2023 Dec 12. mBio. 2024. PMID: 38084997 Free PMC article. - A journey into the brain: insight into how bacterial pathogens cross blood-brain barriers.
Coureuil M, Lécuyer H, Bourdoulous S, Nassif X. Coureuil M, et al. Nat Rev Microbiol. 2017 Mar;15(3):149-159. doi: 10.1038/nrmicro.2016.178. Epub 2017 Jan 16. Nat Rev Microbiol. 2017. PMID: 28090076 Review. - Cytokine Storm-Definition, Causes, and Implications.
Jarczak D, Nierhaus A. Jarczak D, et al. Int J Mol Sci. 2022 Oct 3;23(19):11740. doi: 10.3390/ijms231911740. Int J Mol Sci. 2022. PMID: 36233040 Free PMC article. Review.
References
- Yazdankhah SP, Caugant DA. Neisseria meningitidis: an overview of the carriage state. J. Med. Microbiol. 2004;53:821–832. - PubMed
- Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–2210. - PubMed
- Virji M. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat. Rev. Microbiol. 2009;7:274–286. - PubMed
- Brandtzaeg P, van Deuren M. Classification and pathogenesis of meningococcal infections. Methods Mol. Biol. 2012;799:21–35. - PubMed
- Lemichez E, Lecuit M, Nassif X, Bourdoulous S. Breaking the wall: targeting of the endothelium by pathogenic bacteria. Nat. Rev. Microbiol. 2010;8:93–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases