Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity - PubMed (original) (raw)
Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity
Mart Krupovic et al. BMC Biol. 2014.
Abstract
Background: Diverse transposable elements are abundant in genomes of cellular organisms from all three domains of life. Although transposons are often regarded as junk DNA, a growing body of evidence indicates that they are behind some of the major evolutionary innovations. With the growth in the number and diversity of sequenced genomes, previously unnoticed mobile elements continue to be discovered.
Results: We describe a new superfamily of archaeal and bacterial mobile elements which we denote casposons because they encode Cas1 endonuclease, a key enzyme of the CRISPR-Cas adaptive immunity systems of archaea and bacteria. The casposons share several features with self-synthesizing eukaryotic DNA transposons of the Polinton/Maverick class, including terminal inverted repeats and genes for B family DNA polymerases. However, unlike any other known mobile elements, the casposons are predicted to rely on Cas1 for integration and excision, via a mechanism similar to the integration of new spacers into CRISPR loci. We identify three distinct families of casposons that differ in their gene repertoires and evolutionary provenance of the DNA polymerases. Deep branching of the casposon-encoded endonuclease in the Cas1 phylogeny suggests that casposons played a pivotal role in the emergence of CRISPR-Cas immunity.
Conclusions: The casposons are a novel superfamily of mobile elements, the first family of putative self-synthesizing transposons discovered in prokaryotes. The likely contribution of capsosons to the evolution of CRISPR-Cas parallels the involvement of the RAG1 transposase in vertebrate immunoglobulin gene rearrangement, suggesting that recruitment of endonucleases from mobile elements as ready-made tools for genome manipulation is a general route of evolution of adaptive immunity.
Figures
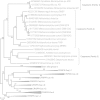
Figure 1
Phylogeny of Cas1 proteins. Cas1 proteins encoded by casposons are represented in the framework of the Cas1 sequences representing the major types of CRISPR-Cas system. All clades including Cas1 from different CRISPR-Cas systems as well as Cas1-solo group 1 were collapsed for clarity (altogether, 52 non-casposon Cas1 sequences were analyzed, a representative subset of a larger collection of Cas1 sequences analyzed previously [33]; the Cas1 sequence alignment used to generate the tree is provided in Additional file 2, whereas the tree in which all branches are expanded is shown in Additional file 1: Figure S5). Numbers at the branch points represent RELL (resampling of estimated log-likelihoods)-like local support values calculated by FastTree.
Figure 2
Casposon genome maps. The three families of casposons are indicated. The precise nucleotide coordinates of depicted casposons can be found in Additional file 1: Table S1. Predicted protein-coding genes are indicated with arrows, indicating the direction of transcription. The color key for the designation of the common genes is shown in the top right area of the figure. ‘HTH (other)’ denotes proteins that are not orthologous but nevertheless contain HTH domains. Terminal inverted repeats (TIR) are shown with black rectangles and their sequences are shown in Additional file 1: Figure S2. The grey boxes outlined with a broken line in MetPsy-C1 depict duplicated regions. The striped green arrows represent genes encoding divergent HNH proteins. Abbreviations: ZBD, zinc-binding domain-containing protein; HNH, HNH family endonuclease; HTH, helix-turn-helix proteins; MTase, methyltransferase; RHH, ribbon-helix-helix protein; REase, restriction endonuclease; AP, apurinic/apyrimidinic; UDG, uracil-DNA glycosylase; SFI, superfamily I; IR, inverted repeat.
Figure 3
Phylogeny of protein-primed B family DNA polymerases. Clades that are only distantly related to the casposon-encoded proteins were collapsed. The tree is rooted with phi29-like bacteriophages of the Podoviridae family. Numbers at the branch points represent RELL (resampling of estimated log-likelihoods)-like local support values calculated by FastTree.
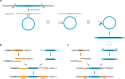
Figure 4
Proposed mechanisms of casposon replication and integration. (a) Mechanism of casposon DNA excision and replication. (b) Mechanism of spacer acquisition by CRISPR-Cas system. Adapted from [34]. (c) Proposed mechanism of Cas1-mediated casposon integration. See text for details. Subterminal regions within TIRs outlined with the broken line indicate that processing occurs only in a fraction of casposons. See text for details. Abbreviations: TIR, terminal inverted repeats; TSD, target site duplication.
Similar articles
- Recent Mobility of Casposons, Self-Synthesizing Transposons at the Origin of the CRISPR-Cas Immunity.
Krupovic M, Shmakov S, Makarova KS, Forterre P, Koonin EV. Krupovic M, et al. Genome Biol Evol. 2016 Jan 13;8(2):375-86. doi: 10.1093/gbe/evw006. Genome Biol Evol. 2016. PMID: 26764427 Free PMC article. - Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems.
Krupovic M, Koonin EV. Krupovic M, et al. Curr Opin Microbiol. 2016 Jun;31:25-33. doi: 10.1016/j.mib.2016.01.006. Epub 2016 Feb 1. Curr Opin Microbiol. 2016. PMID: 26836982 Free PMC article. Review. - Casposons - silent heroes of the CRISPR-Cas systems evolutionary history.
Smaruj P, Kieliszek M. Smaruj P, et al. EXCLI J. 2023 Jan 5;22:70-83. doi: 10.17179/excli2022-5581. eCollection 2023. EXCLI J. 2023. PMID: 36814855 Free PMC article. Review. - Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back.
Koonin EV, Makarova KS. Koonin EV, et al. Genome Biol Evol. 2017 Oct 1;9(10):2812-2825. doi: 10.1093/gbe/evx192. Genome Biol Evol. 2017. PMID: 28985291 Free PMC article. Review. - CRISPR-Cas immunity and mobile DNA: a new superfamily of DNA transposons encoding a Cas1 endonuclease.
Hickman AB, Dyda F. Hickman AB, et al. Mob DNA. 2014 Aug 26;5:23. doi: 10.1186/1759-8753-5-23. eCollection 2014. Mob DNA. 2014. PMID: 25180049 Free PMC article.
Cited by
- A bend, flip and trap mechanism for transposon integration.
Morris ER, Grey H, McKenzie G, Jones AC, Richardson JM. Morris ER, et al. Elife. 2016 May 25;5:e15537. doi: 10.7554/eLife.15537. Elife. 2016. PMID: 27223327 Free PMC article. - Structural biology of CRISPR-Cas immunity and genome editing enzymes.
Wang JY, Pausch P, Doudna JA. Wang JY, et al. Nat Rev Microbiol. 2022 Nov;20(11):641-656. doi: 10.1038/s41579-022-00739-4. Epub 2022 May 13. Nat Rev Microbiol. 2022. PMID: 35562427 Review. - Evolutionary plasticity and functional versatility of CRISPR systems.
Koonin EV, Makarova KS. Koonin EV, et al. PLoS Biol. 2022 Jan 5;20(1):e3001481. doi: 10.1371/journal.pbio.3001481. eCollection 2022 Jan. PLoS Biol. 2022. PMID: 34986140 Free PMC article. - Evolution of Immune Systems From Viruses and Transposable Elements.
Broecker F, Moelling K. Broecker F, et al. Front Microbiol. 2019 Jan 29;10:51. doi: 10.3389/fmicb.2019.00051. eCollection 2019. Front Microbiol. 2019. PMID: 30761103 Free PMC article. Review. - Mixed waste contamination selects for a mobile genetic element population enriched in multiple heavy metal resistance genes.
Goff JL, Lui LM, Nielsen TN, Poole FL, Smith HJ, Walker KF, Hazen TC, Fields MW, Arkin AP, Adams MWW. Goff JL, et al. ISME Commun. 2024 May 9;4(1):ycae064. doi: 10.1093/ismeco/ycae064. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38800128 Free PMC article.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. et al.Initial sequencing and analysis of the human genome. Nature. 2001;12:860–921. doi: 10.1038/35057062. - DOI - PubMed
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N. et al.The sequence of the human genome. Science. 2001;12:1304–1351. doi: 10.1126/science.1058040. - DOI - PubMed
- López-Flores I, Garrido-Ramos MA. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 2012;12:1–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources