RNA-seq: impact of RNA degradation on transcript quantification - PubMed (original) (raw)
RNA-seq: impact of RNA degradation on transcript quantification
Irene Gallego Romero et al. BMC Biol. 2014.
Abstract
Background: The use of low quality RNA samples in whole-genome gene expression profiling remains controversial. It is unclear if transcript degradation in low quality RNA samples occurs uniformly, in which case the effects of degradation can be corrected via data normalization, or whether different transcripts are degraded at different rates, potentially biasing measurements of expression levels. This concern has rendered the use of low quality RNA samples in whole-genome expression profiling problematic. Yet, low quality samples (for example, samples collected in the course of fieldwork) are at times the sole means of addressing specific questions.
Results: We sought to quantify the impact of variation in RNA quality on estimates of gene expression levels based on RNA-seq data. To do so, we collected expression data from tissue samples that were allowed to decay for varying amounts of time prior to RNA extraction. The RNA samples we collected spanned the entire range of RNA Integrity Number (RIN) values (a metric commonly used to assess RNA quality). We observed widespread effects of RNA quality on measurements of gene expression levels, as well as a slight but significant loss of library complexity in more degraded samples.
Conclusions: While standard normalizations failed to account for the effects of degradation, we found that by explicitly controlling for the effects of RIN using a linear model framework we can correct for the majority of these effects. We conclude that in instances in which RIN and the effect of interest are not associated, this approach can help recover biologically meaningful signals in data from degraded RNA samples.
Figures
Figure 1
Broad effects of RNA degradation. A) PCA plot of the 15 samples included in the study based on data from 29,156 genes with at least one mapped read in a single individual. Different colors identify different time-points, while each shape indicates a particular individual in the data set. B) Spearman correlation plot of the 15 samples in the study. PCA, principal component analysis.
Figure 2
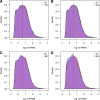
Changes in library complexity over time. Dashed lines indicate median RPKM at each time-point. A) Density plots of RPKM values among all three individuals at 0 hours and 12 hours. B) as A, but 0 hours and 24 hours. C) as A, but 0 hours and 48 hours. D) as A, but 0 hours and 84 hours. RPKM, reads per kilobase transcript per million.
Figure 3
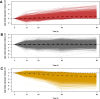
Log 10 median abundance of genes across all three individuals relative to 0 hours. Plots are separated by slope. A) Transcripts with significantly slow rates of degradation relative to the mean rate (identified at 1% FDR, n = 3,745). B) Transcripts that are degraded at a rate close to the mean cellular rate (n = 4,656). C) Transcripts with significantly fast rates of degradation relative to the mean rate (identified at 1% FDR, n = 3,522). In all plots, the thick dashed line indicates the median degradation rate for all genes in that group, whereas the thin dashed line denotes no change in degradation rate relative to 0 hours. FDR, false discovery rate.
Figure 4
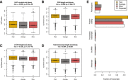
Characteristics of rapidly and slowly degraded transcripts. In all plots, rapidly degraded transcripts are plotted in gold, transcripts degraded at an average rate are plotted in grey and slowly degraded transcripts are in red. A) By transcript %GC content. B) By coding region length. C) By 3′UTR length. D) By complete transcript length. E) By ENSEMBL biotype.
Figure 5
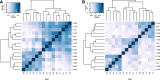
Spearman correlation matrices of the top 10% genes with high inter-individual variance at 0 hours. A) Before RIN correction. B) After regressing the effects of RIN. RIN, RNA integrity number.
Similar articles
- Effects of RNA integrity on transcript quantification by total RNA sequencing of clinically collected human placental samples.
Reiman M, Laan M, Rull K, Sõber S. Reiman M, et al. FASEB J. 2017 Aug;31(8):3298-3308. doi: 10.1096/fj.201601031RR. Epub 2017 Apr 26. FASEB J. 2017. PMID: 28446590 - Measure transcript integrity using RNA-seq data.
Wang L, Nie J, Sicotte H, Li Y, Eckel-Passow JE, Dasari S, Vedell PT, Barman P, Wang L, Weinshiboum R, Jen J, Huang H, Kohli M, Kocher JP. Wang L, et al. BMC Bioinformatics. 2016 Feb 3;17:58. doi: 10.1186/s12859-016-0922-z. BMC Bioinformatics. 2016. PMID: 26842848 Free PMC article. - Sequencing degraded RNA addressed by 3' tag counting.
Sigurgeirsson B, Emanuelsson O, Lundeberg J. Sigurgeirsson B, et al. PLoS One. 2014 Mar 14;9(3):e91851. doi: 10.1371/journal.pone.0091851. eCollection 2014. PLoS One. 2014. PMID: 24632678 Free PMC article. - A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples.
Schuierer S, Carbone W, Knehr J, Petitjean V, Fernandez A, Sultan M, Roma G. Schuierer S, et al. BMC Genomics. 2017 Jun 5;18(1):442. doi: 10.1186/s12864-017-3827-y. BMC Genomics. 2017. PMID: 28583074 Free PMC article. - Impact of RNA degradation on fusion detection by RNA-seq.
Davila JI, Fadra NM, Wang X, McDonald AM, Nair AA, Crusan BR, Wu X, Blommel JH, Jen J, Rumilla KM, Jenkins RB, Aypar U, Klee EW, Kipp BR, Halling KC. Davila JI, et al. BMC Genomics. 2016 Oct 20;17(1):814. doi: 10.1186/s12864-016-3161-9. BMC Genomics. 2016. PMID: 27765019 Free PMC article.
Cited by
- Gene Expression Profiling in the Hibernating Primate, Cheirogaleus Medius.
Faherty SL, Villanueva-Cañas JL, Klopfer PH, Albà MM, Yoder AD. Faherty SL, et al. Genome Biol Evol. 2016 Aug 25;8(8):2413-26. doi: 10.1093/gbe/evw163. Genome Biol Evol. 2016. PMID: 27412611 Free PMC article. - Robust Acquisition of Spatial Transcriptional Programs in Tissues With Immunofluorescence-Guided Laser Capture Microdissection.
Zhang X, Hu C, Huang C, Wei Y, Li X, Hu M, Li H, Wu J, Czajkowsky DM, Guo Y, Shao Z. Zhang X, et al. Front Cell Dev Biol. 2022 Mar 25;10:853188. doi: 10.3389/fcell.2022.853188. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35399504 Free PMC article. - Gene expression profiling of whole blood: A comparative assessment of RNA-stabilizing collection methods.
Donohue DE, Gautam A, Miller SA, Srinivasan S, Abu-Amara D, Campbell R, Marmar CR, Hammamieh R, Jett M. Donohue DE, et al. PLoS One. 2019 Oct 10;14(10):e0223065. doi: 10.1371/journal.pone.0223065. eCollection 2019. PLoS One. 2019. PMID: 31600258 Free PMC article. - Molecular Classification and Interpretation of Amyotrophic Lateral Sclerosis Using Deep Convolution Neural Networks and Shapley Values.
Karim A, Su Z, West PK, Keon M, The Nygc Als Consortium, Shamsani J, Brennan S, Wong T, Milicevic O, Teunisse G, Rad HN, Sattar A. Karim A, et al. Genes (Basel). 2021 Oct 30;12(11):1754. doi: 10.3390/genes12111754. Genes (Basel). 2021. PMID: 34828360 Free PMC article. - A method for simultaneous detection of small and long RNA biotypes by ribodepleted RNA-Seq.
Potemkin N, Cawood SMF, Treece J, Guévremont D, Rand CJ, McLean C, Stanton JL, Williams JM. Potemkin N, et al. Sci Rep. 2022 Jan 12;12(1):621. doi: 10.1038/s41598-021-04209-4. Sci Rep. 2022. PMID: 35022475 Free PMC article.
References
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;12:436–442. doi: 10.1038/nbt.1861. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous