Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant Jatropha curcas - PubMed (original) (raw)
Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant Jatropha curcas
Chaoqiong Li et al. BMC Plant Biol. 2014.
Abstract
Background: Physic nut (Jatropha curcas L.) is a potential feedstock for biofuel production because Jatropha oil is highly suitable for the production of the biodiesel and bio-jet fuels. However, Jatropha exhibits low seed yield as a result of unreliable and poor flowering. FLOWERING LOCUS T (FT) -like genes are important flowering regulators in higher plants. To date, the flowering genes in Jatropha have not yet been identified or characterized.
Results: To better understand the genetic control of flowering in Jatropha, an FT homolog was isolated from Jatropha and designated as JcFT. Sequence analysis and phylogenetic relationship of JcFT revealed a high sequence similarity with the FT genes of Litchi chinensis, Populus nigra and other perennial plants. JcFT was expressed in all tissues of adult plants except young leaves, with the highest expression level in female flowers. Overexpression of JcFT in Arabidopsis and Jatropha using the constitutive promoter cauliflower mosaic virus 35S or the phloem-specific promoter Arabidopsis SUCROSE TRANSPORTER 2 promoter resulted in an extremely early flowering phenotype. Furthermore, several flowering genes downstream of JcFT were up-regulated in the JcFT-overexpression transgenic plant lines.
Conclusions: JcFT may encode a florigen that acts as a key regulator in flowering pathway. This study is the first to functionally characterize a flowering gene, namely, JcFT, in the biofuel plant Jatropha.
Figures
Figure 1
Comparison of JcFT and other FT -like genes. (A) Gene structures of JcFT, Hd3a, and AtFT. Boxes indicate exons and thin lines indicate introns. Exon sizes are indicated above each box. (B) Sequence alignment of amino acid sequences. Identical amino acid residues are shaded in black, and similar residues are shaded in gray. Dots denote gaps. Boxes indicating the 14-amino-acid stretch (segment B) and the LYN triad (segment C), and "Y" indicating the highly conserved amino acid Tyr (Y).
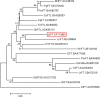
Figure 2
Phylogenetic analysis of the FT homologs from different plant species. Species abbreviations: At, Arabidopsis thaliana; Ci, Citrus unshiu; Cp, Carica papaya; Cs, Cucumis sativus; Fc, Ficus carica; Gh, Gossypium hirsutum; Gt, Gentiana triflora; Jc, Jatropha curcus; Lc, Litchi chinensis; Lt, Lolium temulentum; Md, Malus domestica; Os, Oryza sativa; Phm, Phyllostachys meyeri; Pm, Prunus mume; Pn, Populus nigra; Pp, Prunus persica; Rc, Rosa chinensis; Sl, Solanum lycopersicum; Ta, Triticum aestivum.
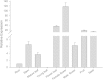
Figure 3
Expression of JcFT in various organs of three-year-old adult Jatropha . The qRT-PCR results were obtained from two independent biological replicates and three technical replicates for each sample. The levels of detected amplicons were normalized using the amplified products of the JcActin1. The mRNA level in the root tissue was set as the standard with a value of 1.
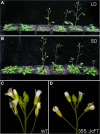
Figure 4
Ectopic expression of JcFT causes early flowering in transgenic Arabidopsis. Growth under LD conditions (A) and SD conditions (B) at 28 days and 45 days after germination, respectively. Left to right: WT, ft-10, 35S::JcFT in Col, SUC2::JcFT in Col, 35S::JcFT in ft-10, and SUC2::JcFT in ft-10. (C and D) Inflorescences of WT and 35S::JcFT transgenic plants.
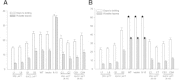
Figure 5
Ectopic expression of JcFT affects flowering in Arabidopsis . (A) Days and leaves to bolting for several JcFT overexpression (CaMV 35S) and phloem- specific expression (SUC2) transgenic Arabidopsis lines, empty vector-transformed plants, WT and mutant ft-10 plants grown under LD conditions. (B) Days and leaves to bolting for transgenic lines in the Col and ft-10 background grown under SD conditions. Values are means ± SD of the results from ten plants of each transgenic line. Arrows at the top of bars for WT, empty vector-transformed Col and ft-10 indicate that plants have not flowered.
Figure 6
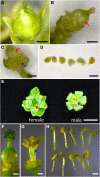
Early flowering of 35S:: JcFT transgenic Jatropha cultured in vitro. (A and B) Flower buds of transgenic Jatropha cultured in vitro for seven weeks. (C) Inflorescence of transgenic Jatropha cultured in vitro. (D) Inflorescence of wild Jatropha in the field. Red arrows indicate flower buds.
Figure 7
Abnormal flowers of transgenic Jatropha harboring 35S:: JcFT . (A) A female flower of transgenic Jatropha cultured in vitro. (B) Pistil of a transgenic female flower. (C) An abnormal hermaphrodite flower of transgenic Jatropha cultured in vitro. (D) Abnormal stamens from an abnormal hermaphrodite flower of transgenic Jatropha shown in (C). (E) Normal female and male flowers of wild Jatropha grown in the field. (F) Pistil of a wild-type female flower. (G and H) Stamens of a wild-type male flower. Bars in (A)-(D) and (F)-(H) represent 1 mm, and bar in (E) represents 5 mm. Red arrows indicate pistils, and blue arrows indicate stamens.
Figure 8
Quantitative RT-PCR analysis of flowering genes downstream of JcFT in WT and 35S:: JcFT transgenic Jatropha . The qRT-PCR results were obtained using two independent biological replicates and three technical replicates for each RNA sample extracted from apex of the 35S::JcFT transgenic and wild-type (WT) shoots cultured in vitro. Transcript levels were normalized using JcActin1 gene as a reference. The mRNA level in WT was set as the standard with a value of 1.
Similar articles
- Ectopic expression of Jatropha curcas APETALA1 (JcAP1) caused early flowering in Arabidopsis, but not in Jatropha.
Tang M, Tao YB, Xu ZF. Tang M, et al. PeerJ. 2016 Apr 25;4:e1969. doi: 10.7717/peerj.1969. eCollection 2016. PeerJ. 2016. PMID: 27168978 Free PMC article. - Efficiency of graft-transmitted JcFT for floral induction in woody perennial species of the Jatropha genus depends on transport distance.
Tang M, Bai X, Wang J, Chen T, Meng X, Deng H, Li C, Xu ZF. Tang M, et al. Tree Physiol. 2022 Jan 5;42(1):189-201. doi: 10.1093/treephys/tpab116. Tree Physiol. 2022. PMID: 34505154 Free PMC article. - Inhibition of flowering by gibberellins in the woody plant Jatropha curcas is restored by overexpression of JcFT.
Huang P, Yang J, Ke J, Cai L, Hu Y, Ni J, Li C, Xu ZF, Tang M. Huang P, et al. Plant Sci. 2024 Jul;344:112100. doi: 10.1016/j.plantsci.2024.112100. Epub 2024 Apr 26. Plant Sci. 2024. PMID: 38679393 - Promotion of natural flowers by JcFT depends on JcLFY in the perennial woody species Jatropha curcas.
Bai X, Ke J, Huang P, Fatima I, Cheng T, Tang M. Bai X, et al. Plant Sci. 2022 May;318:111236. doi: 10.1016/j.plantsci.2022.111236. Epub 2022 Feb 24. Plant Sci. 2022. PMID: 35351308 - Three TFL1 homologues regulate floral initiation in the biofuel plant Jatropha curcas.
Li C, Fu Q, Niu L, Luo L, Chen J, Xu ZF. Li C, et al. Sci Rep. 2017 Feb 22;7:43090. doi: 10.1038/srep43090. Sci Rep. 2017. PMID: 28225036 Free PMC article.
Cited by
- Molecular Mechanisms of the Floral Biology of _Jatropha curca_s: Opportunities and Challenges as an Energy Crop.
Gangwar M, Shankar J. Gangwar M, et al. Front Plant Sci. 2020 Jun 9;11:609. doi: 10.3389/fpls.2020.00609. eCollection 2020. Front Plant Sci. 2020. PMID: 32582231 Free PMC article. Review. - An ortholog of the MADS-box gene SEPALLATA3 regulates stamen development in the woody plant Jatropha curcas.
Zhao ML, Zhou ZF, Chen MS, Xu CJ, Xu ZF. Zhao ML, et al. Planta. 2022 Apr 27;255(6):111. doi: 10.1007/s00425-022-03886-3. Planta. 2022. PMID: 35478059 - An efficient protocol for _Agrobacterium_-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection.
Fu Q, Li C, Tang M, Tao YB, Pan BZ, Zhang L, Niu L, He H, Wang X, Xu ZF. Fu Q, et al. Plant Biotechnol Rep. 2015;9(6):405-416. doi: 10.1007/s11816-015-0377-0. Epub 2015 Nov 6. Plant Biotechnol Rep. 2015. PMID: 26640597 Free PMC article. - Ectopic expression of Jatropha curcas APETALA1 (JcAP1) caused early flowering in Arabidopsis, but not in Jatropha.
Tang M, Tao YB, Xu ZF. Tang M, et al. PeerJ. 2016 Apr 25;4:e1969. doi: 10.7717/peerj.1969. eCollection 2016. PeerJ. 2016. PMID: 27168978 Free PMC article. - Gibberellin Promotes Shoot Branching in the Perennial Woody Plant Jatropha curcas.
Ni J, Gao C, Chen MS, Pan BZ, Ye K, Xu ZF. Ni J, et al. Plant Cell Physiol. 2015 Aug;56(8):1655-66. doi: 10.1093/pcp/pcv089. Epub 2015 Jun 15. Plant Cell Physiol. 2015. PMID: 26076970 Free PMC article.
References
- Divakara B, Upadhyaya H, Wani S, Gowda C. Biology and genetic improvement of Jatropha curcas L.: A review. Appl Energy. 2010;87:732–742. doi: 10.1016/j.apenergy.2009.07.013. - DOI
- Akashi K. Jatropha research: A new frontier for biofuel development. Plant Biotechnol. 2012;29:121. doi: 10.5511/plantbiotechnology.12.0003p. - DOI
- Khalil H, Aprilia N, Bhat A, Jawaid M, Paridah M, Rudi D. A Jatropha biomass as renewable materials for biocomposites and its applications. Renew Sust Energ Rev. 2013;22:667–685.
- Pandey VC, Singh K, Singh JS, Kumar A, Singh B, Singh RP. Jatropha curcas: A potential biofuel plant for sustainable environmental development. Renew Sust Energy Rev. 2012;16:2870–2883. doi: 10.1016/j.rser.2012.02.004. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources