Evaluation and Structural Basis for the Inhibition of Tankyrases by PARP Inhibitors - PubMed (original) (raw)
. 2013 Nov 20;5(1):18-22.
doi: 10.1021/ml400292s. eCollection 2014 Jan 9.
Affiliations
- PMID: 24900770
- PMCID: PMC4027629
- DOI: 10.1021/ml400292s
Evaluation and Structural Basis for the Inhibition of Tankyrases by PARP Inhibitors
Teemu Haikarainen et al. ACS Med Chem Lett. 2013.
Abstract
Tankyrases, an enzyme subfamily of human poly(ADP-ribosyl)polymerases, are potential drug targets especially against cancer. We have evaluated inhibition of tankyrases by known PARP inhibitors and report five cocrystal structures of the most potent compounds in complex with human tankyrase 2. The inhibitors include the small general PARP inhibitors Phenanthridinone, PJ-34, and TIQ-A as well as the more advanced inhibitors EB-47 and rucaparib. The compounds anchor to the nicotinamide subsite of tankyrase 2. Crystal structures reveal flexibility of the ligand binding site with implications for drug development against tankyrases and other ADP-ribosyltransferases. EB-47 mimics the substrate NAD(+) and extends from the nicotinamide to the adenosine subsite. The clinical ARTD1 inhibitor candidate rucaparib was the most potent tankyrase inhibitor identified (24 and 14 nM for tankyrases), which indicates that inhibition of tankyrases would affect the cellular responses of this compound.
Keywords: ADP-ribosyltransferase; PARP; Tankyrase; inhibitor; poly(ADP-ribose) polymerase.
Figures
Figure 1
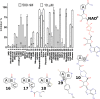
Screening of PARP inhibitor compounds and the structures of the potent hit compounds. At 10 μM many of the compounds inhibited ARTD5, whereas a test at 500 nM identified 9 compounds that showed over 20% inhibition. Data shown are mean ± SD.
Figure 2
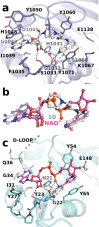
Crystal structures of (a) phenanthridinone (16), (b) TIQ-A (18), (c) PJ-34 (17), and (d) rucaparib (29) bound to the ARTD6 catalytic fragment. The compounds are shown as ball-and-stick models and the hydrogen bonds with the protein are shown as dashed lines. The disordered D-loop in panels c and d is shown as a thick dashed line.
Figure 3
Binding of the dual site inhibitor EB-47 (10) and comparison of the structure with the substrate NAD+ binding mode to Diphtheria toxin. (a) Binding mode of EB-47 to ARTD6. (b) Comparison of superposed EB-47 with NAD+ bound to Diphtheria toxin (PDB code 1TOX). Superposition was done with selected atoms in order to overlap adenosine and nicotinamide moieties. (c) NAD+ complex conformation observed in Diphtheria toxin (PDB code 1TOX). The disordered D-loop is shown as a dashed line.
Similar articles
- Tankyrases as drug targets.
Lehtiö L, Chi NW, Krauss S. Lehtiö L, et al. FEBS J. 2013 Aug;280(15):3576-93. doi: 10.1111/febs.12320. Epub 2013 Jun 18. FEBS J. 2013. PMID: 23648170 Review. - Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
Thorsell AG, Ekblad T, Karlberg T, Löw M, Pinto AF, Trésaugues L, Moche M, Cohen MS, Schüler H. Thorsell AG, et al. J Med Chem. 2017 Feb 23;60(4):1262-1271. doi: 10.1021/acs.jmedchem.6b00990. Epub 2016 Dec 21. J Med Chem. 2017. PMID: 28001384 Free PMC article. - Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell Å, Ekblad T, Öncü D, Kull B, Robertson GM, Pellicciari R, Schüler H, Weigelt J. Wahlberg E, et al. Nat Biotechnol. 2012 Feb 19;30(3):283-8. doi: 10.1038/nbt.2121. Nat Biotechnol. 2012. PMID: 22343925 - Structural basis of selective inhibition of human tankyrases.
Narwal M, Venkannagari H, Lehtiö L. Narwal M, et al. J Med Chem. 2012 Feb 9;55(3):1360-7. doi: 10.1021/jm201510p. Epub 2012 Jan 25. J Med Chem. 2012. PMID: 22233320 - Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naïve pluripotency.
Zimmerlin L, Zambidis ET. Zimmerlin L, et al. Exp Cell Res. 2020 May 1;390(1):111935. doi: 10.1016/j.yexcr.2020.111935. Epub 2020 Mar 7. Exp Cell Res. 2020. PMID: 32151493 Free PMC article. Review.
Cited by
- ClickGen: Directed exploration of synthesizable chemical space via modular reactions and reinforcement learning.
Wang M, Li S, Wang J, Zhang O, Du H, Jiang D, Wu Z, Deng Y, Kang Y, Pan P, Li D, Wang X, Yao X, Hou T, Hsieh CY. Wang M, et al. Nat Commun. 2024 Nov 22;15(1):10127. doi: 10.1038/s41467-024-54456-y. Nat Commun. 2024. PMID: 39578485 Free PMC article. - PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Dawicki-McKenna JM, Langelier MF, DeNizio JE, Riccio AA, Cao CD, Karch KR, McCauley M, Steffen JD, Black BE, Pascal JM. Dawicki-McKenna JM, et al. Mol Cell. 2015 Dec 3;60(5):755-768. doi: 10.1016/j.molcel.2015.10.013. Epub 2015 Nov 25. Mol Cell. 2015. PMID: 26626480 Free PMC article. - PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes.
Gupte R, Liu Z, Kraus WL. Gupte R, et al. Genes Dev. 2017 Jan 15;31(2):101-126. doi: 10.1101/gad.291518.116. Genes Dev. 2017. PMID: 28202539 Free PMC article. Review. - Exclusive destruction of mitotic spindles in human cancer cells.
Visochek L, Castiel A, Mittelman L, Elkin M, Atias D, Golan T, Izraeli S, Peretz T, Cohen-Armon M. Visochek L, et al. Oncotarget. 2017 Mar 28;8(13):20813-20824. doi: 10.18632/oncotarget.15343. Oncotarget. 2017. PMID: 28209915 Free PMC article. - Structural basis of tankyrase activation by polymerization.
Pillay N, Mariotti L, Zaleska M, Inian O, Jessop M, Hibbs S, Desfosses A, Hopkins PCR, Templeton CM, Beuron F, Morris EP, Guettler S. Pillay N, et al. Nature. 2022 Dec;612(7938):162-169. doi: 10.1038/s41586-022-05449-8. Epub 2022 Nov 23. Nature. 2022. PMID: 36418402 Free PMC article.
References
- Bryant H. E.; Schultz N.; Thomas H. D.; Parker K. M.; Flower D.; Lopez E.; Kyle S.; Meuth M.; Curtin N. J.; Helleday T. Specific Killing of BRCA2-Deficient Tumours with Inhibitors of Poly(ADP-ribose) Polymerase. Nature 2005, 434, 913–917. - PubMed
- Riffell J. L.; Lord C. J.; Ashworth A. Tankyrase-Targeted Therapeutics: Expanding Opportunities in the PARP Family. Nat. Rev. Drug Discovery 2012, 11, 923–936. - PubMed
- Lehtiö L.; Chi N.-W.; Krauss S. Tankyrases as Drug Targets. FEBS J. 2013, 280, 3576–3593. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information