Reprogramming the methylome: erasing memory and creating diversity - PubMed (original) (raw)
Reprogramming the methylome: erasing memory and creating diversity
Heather J Lee et al. Cell Stem Cell. 2014.
Abstract
The inheritance of epigenetic marks, in particular DNA methylation, provides a molecular memory that ensures faithful commitment to transcriptional programs during mammalian development. Epigenetic reprogramming results in global hypomethylation of the genome together with a profound loss of memory, which underlies naive pluripotency. Such global reprogramming occurs in primordial germ cells, early embryos, and embryonic stem cells where reciprocal molecular links connect the methylation machinery to pluripotency. Priming for differentiation is initiated upon exit from pluripotency, and we propose that epigenetic mechanisms create diversity of transcriptional states, which help with symmetry breaking during cell fate decisions and lineage commitment.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
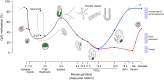
Global CpG Methylation Levels in the Mouse Germline, Somatic Tissues, and ESCs The mouse germline undergoes two major waves of demethylation, the first in the early embryo where the paternal genome (blue) is actively demethylated prior to and during replication. Both the paternal and maternal (red) genomes passively lose methylation after this until the blastocyst stage (E3.5). The second wave of demethylation occurs in the primordial germ cells between E6.5 and E13.5 as they emerge from the epiblast. Methylation is then re-established in a sex-specific manner after E13.5 and the nongrowing (NG) oocyte stage, in males and females, respectively, eventually giving rise to mature gametic patterns. Naive and primed ESCs can be cultured from the ICM or be interchanged with each other (dashed line), by growth in either serum or 2i media, respectively. Only naive ESCs display low methylation (∼30%) that corresponds to in vivo pluripotent tissues (shaded boxes on the far right). Erased cells display less than 10% methylation, whereas somatic tissues (derived from the E6.5 epiblast) show consistently high methylation around 70%–85%. The placenta is relatively demethylated compared to somatic tissues and is derived from the blastocyst trophectoderm (E3.5). In order to compare between genome-wide (Ficz et al., 2013; Hon et al., 2013; Kobayashi et al., 2012; Seisenberger et al., 2012; Shirane et al., 2013) and reduced representation bisulfite sequencing data sets (Smallwood et al., 2011; Smith et al., 2012), 100 kb probes not overlapping CpG islands were analyzed as previously (Ficz et al., 2013).
Figure 2
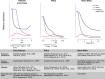
Kinetics of Genome-wide Demethylation in Early Embryos, PGCs, and ESCs Impairment of methylation establishment and maintenance contributes to genome-wide demethylation in vivo (early embryo and PGCs) and in culture (naive ESCs), as do the oxidation and base excision repair pathways. Approximate levels of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are represented by blue and red lines, respectively. Dashed blue lines indicate the expected level of 5mC if demethylation was caused solely by complete inactivation of maintenance methylation.
Figure 3
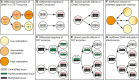
Potential Sources of DNA Methylation Heterogeneity (A) Differential expression of TET or DNMT3 enzymes between cells would lead to global changes in DNA methylation levels. (B and C) Differential recruitment of DNMT3 (B) or TET (C) enzymes at certain loci could generate cells with distinct patterns of DNA methylation. In the case of TET (C) hemi-5hmC would be an intermediate to loss of DNA methylation since DNMT1 does not maintain this mark. (D and E) Strand-specific effects of DNMT3 (D) or TET (E) enzymes could also produce daughter cells with distinct methylation patterns. In each case, hemimodified DNA would be a transitional state. (F) Erasure of inherited methylation patterns (e.g., removal of oocyte derived methylation in the ICM) could also be inefficient and stochastic, generating sister cells with distinct patterns of inherited DNA methylation. (G) Inefficient maintenance of DNA methylation could also produce DNA methylation heterogeneity via hemimethylated intermediaries.
Figure 4
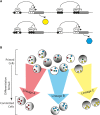
Consequences of DNA Methylation Heterogeneity for Cell Fate Decisions (A) Heterogeneous methylation at regulatory elements (e.g., enhancers and promoters) may affect the binding of transcription factors (TFs) and methyl-binding proteins (MBPs, e.g., MeCP2 and MBD1), that can in turn activate, or repress, gene expression. Black and white circles represent methylated and unmethylated sites, respectively. (B) The result is a pool of diverse cells at the exit from pluripotency in which heterogeneous patterns of methylation (black shading) underlies heterogeneous transcriptional programmes (colored shapes). This cell diversity may predispose cells toward different lineage choices upon receipt of differentiation stimuli.
Similar articles
- Genome-Scale Oscillations in DNA Methylation during Exit from Pluripotency.
Rulands S, Lee HJ, Clark SJ, Angermueller C, Smallwood SA, Krueger F, Mohammed H, Dean W, Nichols J, Rugg-Gunn P, Kelsey G, Stegle O, Simons BD, Reik W. Rulands S, et al. Cell Syst. 2018 Jul 25;7(1):63-76.e12. doi: 10.1016/j.cels.2018.06.012. Cell Syst. 2018. PMID: 30031774 Free PMC article. - What an epigenome remembers.
Lange UC, Schneider R. Lange UC, et al. Bioessays. 2010 Aug;32(8):659-68. doi: 10.1002/bies.201000030. Bioessays. 2010. PMID: 20658704 Review. - Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health.
Xavier MJ, Roman SD, Aitken RJ, Nixon B. Xavier MJ, et al. Hum Reprod Update. 2019 Sep 11;25(5):518-540. doi: 10.1093/humupd/dmz017. Hum Reprod Update. 2019. PMID: 31374565 - Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification.
von Meyenn F, Berrens RV, Andrews S, Santos F, Collier AJ, Krueger F, Osorno R, Dean W, Rugg-Gunn PJ, Reik W. von Meyenn F, et al. Dev Cell. 2016 Oct 10;39(1):104-115. doi: 10.1016/j.devcel.2016.09.015. Dev Cell. 2016. PMID: 27728778 Free PMC article. - Epigenetic reprogramming in mammals.
Morgan HD, Santos F, Green K, Dean W, Reik W. Morgan HD, et al. Hum Mol Genet. 2005 Apr 15;14 Spec No 1:R47-58. doi: 10.1093/hmg/ddi114. Hum Mol Genet. 2005. PMID: 15809273 Review.
Cited by
- Structure and Function of TET Enzymes.
Yin X, Hu L, Xu Y. Yin X, et al. Adv Exp Med Biol. 2022;1389:239-267. doi: 10.1007/978-3-031-11454-0_10. Adv Exp Med Biol. 2022. PMID: 36350513 - Function and information content of DNA methylation.
Schübeler D. Schübeler D. Nature. 2015 Jan 15;517(7534):321-6. doi: 10.1038/nature14192. Nature. 2015. PMID: 25592537 Review. - A germ cell-specific ageing pattern in otherwise healthy men.
Laurentino S, Cremers JF, Horsthemke B, Tüttelmann F, Czeloth K, Zitzmann M, Pohl E, Rahmann S, Schröder C, Berres S, Redmann K, Krallmann C, Schlatt S, Kliesch S, Gromoll J. Laurentino S, et al. Aging Cell. 2020 Oct;19(10):e13242. doi: 10.1111/acel.13242. Epub 2020 Sep 20. Aging Cell. 2020. PMID: 32951333 Free PMC article. - Rebooting the Epigenomes during Mammalian Early Embryogenesis.
Xia W, Xie W. Xia W, et al. Stem Cell Reports. 2020 Dec 8;15(6):1158-1175. doi: 10.1016/j.stemcr.2020.09.005. Epub 2020 Oct 8. Stem Cell Reports. 2020. PMID: 33035464 Free PMC article. Review. - Wnt Inhibition Facilitates RNA-Mediated Reprogramming of Human Somatic Cells to Naive Pluripotency.
Bredenkamp N, Yang J, Clarke J, Stirparo GG, von Meyenn F, Dietmann S, Baker D, Drummond R, Ren Y, Li D, Wu C, Rostovskaya M, Eminli-Meissner S, Smith A, Guo G. Bredenkamp N, et al. Stem Cell Reports. 2019 Dec 10;13(6):1083-1098. doi: 10.1016/j.stemcr.2019.10.009. Epub 2019 Nov 7. Stem Cell Reports. 2019. PMID: 31708477 Free PMC article.
References
- Beraldi R., Pittoggi C., Sciamanna I., Mattei E., Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 2006;73:279–287. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources