The 3D genome in transcriptional regulation and pluripotency - PubMed (original) (raw)
Review
The 3D genome in transcriptional regulation and pluripotency
David U Gorkin et al. Cell Stem Cell. 2014.
Abstract
It can be convenient to think of the genome as simply a string of nucleotides, the linear order of which encodes an organism's genetic blueprint. However, the genome does not exist as a linear entity within cells where this blueprint is actually utilized. Inside the nucleus, the genome is organized in three-dimensional (3D) space, and lineage-specific transcriptional programs that direct stem cell fate are implemented in this native 3D context. Here, we review principles of 3D genome organization in mammalian cells. We focus on the emerging relationship between genome organization and lineage-specific transcriptional regulation, which we argue are inextricably linked.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Figure 1. Different levels of genome organization
[From top to bottom] Level 1: Chromosomes occupy distinct sub-regions of the nucleus known as chromosome territories (CTs). Individual chromosomes are indicated by different colors. Level 2: Transcriptionally inactive regions are enriched at the nuclear periphery where they contact the nuclear lamina (red). Actively transcribed genes often co-localize at RNA polymerase II transcription factories (yellow). These and other instances of co-localization between regions with similar transcriptional activity may provide the physical basis for the observations of A and B compartments in C-data. Level 3: Topological domains, or Topologically-Associating Domains (TADs) are regions of frequent local interactions separated by boundaries across which interactions are less frequent. CTCF binding sites and other sequence features (TSS, SINEs; not depicted here) are enriched at TAD boundaries. Note that CTCF also binds within TADs. Cohesin is often present at TAD boundaries, although it is not shown here. Level 4: Transcriptional regulation depends on long-range Interactions between _cis_-regulatory elements such as enhancers (light red) and promoters (light yellow). These cis-regulatory interactions are facilitated by proteins including Transcription Factors (“TFs”; blue), co-factors such as Mediator (“Med”; red) and Cohesin (purple ring), and RNA Polymerase II (“Pol II”; yellow).
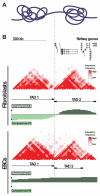
Figure 2. TADs and A/B compartments
A) Diagrammatic representation of two neighboring TADs. B) UCSC genome browser view of the region chr11:115,470,000-116,770,000 which contains two adjacent TADs. [Top] Scale bar and refseq genes. [Middle] C-data from IMR90 fibroblasts. Tracks show pairwise interaction frequencies (red; 40 kb bins), TADs (black bars), and compartments A and B (green). Dashed line marks the boundary between TAD 1 and TAD 2. Note that TAD 2 contains far more genes than TAD 1, and is in the inactive compartment A in IMR90, while TAD 1 is in the inactive compartment A. [Bottom] C-data from human ESCs. Tracks are arranged as above for IMR90. Note that overall TAD structure and location of TAD boundaries do not differ significantly between IMR90 and ESCs. However, TAD1 is in the active compartment A in ESCs. Association of this gene-poor TAD with compartment A in ESCs may be related to the global pervasiveness of open chromatin in pluripotent cells (see section “Genome organization and pluripotency” for further discussion). All C-data taken from Dixon et al. (2012).
Figure 3. Molecular machinery of _cis_-regulatory interactions
[Top] Enhancer-promoter interactions. TFs (blue) bind to enhancers and promoters. RNA polymerase II (yellow) is recruited to the promoter, and Mediator (red) and Cohesin (purple ring) are recruited to the enhancer. Cohesin stabilizes the interaction, perhaps by forming a ring around interacting loci (although there is little experimental evidence to support this at present). [Bottom] ncRNA-mediated interactions. ncRNA-a (orange) have enhancer-like function: they upregulate genes in cis, dependent on Mediator and coincident with 3D interactions between the ncRNA locus and target gene promoter. Involvement of Cohesin in these interactions has not been demonstrated to date.
Similar articles
- Pluripotency in 3D: genome organization in pluripotent cells.
Denholtz M, Plath K. Denholtz M, et al. Curr Opin Cell Biol. 2012 Dec;24(6):793-801. doi: 10.1016/j.ceb.2012.11.001. Epub 2012 Nov 27. Curr Opin Cell Biol. 2012. PMID: 23199754 Free PMC article. Review. - Transcriptional control of pluripotency: decisions in early development.
Johnson BV, Rathjen J, Rathjen PD. Johnson BV, et al. Curr Opin Genet Dev. 2006 Oct;16(5):447-54. doi: 10.1016/j.gde.2006.08.012. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16919449 Review. - Linking Hematopoietic Differentiation to Co-Expressed Sets of Pluripotency-Associated and Imprinted Genes and to Regulatory microRNA-Transcription Factor Motifs.
Hamed M, Trumm J, Spaniol C, Sethi R, Irhimeh MR, Fuellen G, Paulsen M, Helms V. Hamed M, et al. PLoS One. 2017 Jan 4;12(1):e0166852. doi: 10.1371/journal.pone.0166852. eCollection 2017. PLoS One. 2017. PMID: 28052084 Free PMC article. - Distinguishing between mouse and human pluripotent stem cell regulation: the best laid plans of mice and men.
Schnerch A, Cerdan C, Bhatia M. Schnerch A, et al. Stem Cells. 2010 Mar 31;28(3):419-30. doi: 10.1002/stem.298. Stem Cells. 2010. PMID: 20054863 Review.
Cited by
- Giant pandas in captivity undergo short-term adaptation in nerve-related pathways.
Li Y, Xu W, Wang J, Liu H, Liu J, Zhang L, Hou R, Shen F, Liu Y, Cai K. Li Y, et al. BMC Zool. 2024 Feb 21;9(1):4. doi: 10.1186/s40850-024-00195-y. BMC Zool. 2024. PMID: 38383502 Free PMC article. - Spectral identification of topological domains.
Chen J, Hero AO 3rd, Rajapakse I. Chen J, et al. Bioinformatics. 2016 Jul 15;32(14):2151-8. doi: 10.1093/bioinformatics/btw221. Epub 2016 May 5. Bioinformatics. 2016. PMID: 27153657 Free PMC article. - Deconvolution of Ensemble Chromatin Interaction Data Reveals the Latent Mixing Structures in Cell Subpopulations.
Sefer E, Duggal G, Kingsford C. Sefer E, et al. J Comput Biol. 2016 Jun;23(6):425-38. doi: 10.1089/cmb.2015.0210. J Comput Biol. 2016. PMID: 27267775 Free PMC article. - Nuclear Disposition of Alien Chromosome Introgressions into Wheat and Rye Using 3D-FISH.
Koláčková V, Perničková K, Vrána J, Duchoslav M, Jenkins G, Phillips D, Turkosi E, Šamajová O, Sedlářová M, Šamaj J, Doležel J, Kopecký D. Koláčková V, et al. Int J Mol Sci. 2019 Aug 25;20(17):4143. doi: 10.3390/ijms20174143. Int J Mol Sci. 2019. PMID: 31450653 Free PMC article. - Regulation of 3D Organization and Its Role in Cancer Biology.
Peng A, Peng W, Wang R, Zhao H, Yu X, Sun Y. Peng A, et al. Front Cell Dev Biol. 2022 Jun 8;10:879465. doi: 10.3389/fcell.2022.879465. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35757006 Free PMC article. Review.
References
- Akhtar W, De Jong J, Pindyurin AV, Pagie L, Meuleman W, De Ridder J, Berns A, Wessels LF, Van Lohuizen M, Van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–27. - PubMed
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. - PubMed
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, Kharchenko PV, Park PJ, Hochedlinger K. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. - PMC - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. - PubMed
Publication types
MeSH terms
Grants and funding
- U01ES017166/ES/NIEHS NIH HHS/United States
- K12 GM068524/GM/NIGMS NIH HHS/United States
- U01 HL107442/HL/NHLBI NIH HHS/United States
- P50 GM085764/GM/NIGMS NIH HHS/United States
- U54 HG006997/HG/NHGRI NIH HHS/United States
- U01HL107442/HL/NHLBI NIH HHS/United States
- U01 ES017166/ES/NIEHS NIH HHS/United States
- P50GM085764/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources