An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1) - PubMed (original) (raw)
Review
An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1)
Cyril X George et al. J Interferon Cytokine Res. 2014 Jun.
Abstract
Adenosine deaminase acting on RNA1 (ADAR1) catalyzes the C6 deamination of adenosine (A) to produce inosine (I) in regions of RNA with double-stranded (ds) character. This process is known as A-to-I RNA editing. Alternative promoters drive the expression of the Adar1 gene and alternative splicing gives rise to transcripts that encode 2 ADAR1 protein size isoforms. ADAR1 p150 is an interferon (IFN)-inducible dsRNA adenosine deaminase found in the cytoplasm and nucleus, whereas ADAR1 p110 is constitutively expressed and nuclear in localization. Dependent on the duplex structure of the dsRNA substrate, deamination of adenosine by ADAR can be either highly site-selective or nonspecific. A-to-I editing can alter the stability of RNA structures and the coding of RNA as I is read as G instead of A by ribosomes during mRNA translation and by polymerases during RNA replication. A-to-I editing is of broad physiologic significance. Both the production and the action of IFNs, and hence the subsequent interaction of viruses with their hosts, are among the processes affected by A-to-I editing.
Figures
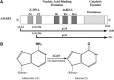
**FIG. 1.
Domain organization and reaction catalyzed by adenosine deaminase acting on RNA1 (ADAR1). (A) Domain organization of human ADAR1. Alternative promoters and alternative splicing generate 2 size-isoforms of ADAR1, an interferon (IFN)-inducible p150 protein and a constitutively expressed p110 protein. The 2 Z-DNA binding domains (Zα, Zβ), the 3 double-stranded RNA (dsRNA) binding domains (RI, RII, and RIII), and the deaminase catalytic domain are shown. Alternative exon 1 and exon 7 structures give rise to the 1,200 amino acid p150 protein and 931 aa p110 protein in human cells. (B) Both p150 and p110 ADAR1 catalyze the hydrolytic C6 deamination of adenosine (A) to yield inosine (I) in dsRNA. Adapted from George and others (2011).
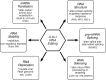
**FIG. 2.
A-to-I RNA editing affects multiple biochemical processes, thereby altering gene expression and function. Because I base pairs as G instead of A, nucleotide substitution of an A with an I may affect mRNA translation by altering a codon potentially leading to an amino acid substitution; RNA structure-dependent activities that trigger IFN responses may be suppressed if dsRNA structures are destabilized; the pre-mRNA splicing pattern of an RNA may be altered by editing a conserved A involved in splice site selection; RNA silencing may be altered by affecting dsRNA structures involved in either micro RNA processing or targeting; RNA virus genome stability may be altered by changing template and therefore complementary product sequences during viral RNA synthesis leading to A-to-G (U-to-C) transitions; and, A-to-I editing of noncoding repetitive (Alu) or nonrepetitive RNA elements may potentially affect RNA stability by altering cellular localization or nuclease recognition. Adapted from Samuel (2011).
Similar articles
- Adenosine deaminases acting on RNA, RNA editing, and interferon action.
George CX, Gan Z, Liu Y, Samuel CE. George CX, et al. J Interferon Cytokine Res. 2011 Jan;31(1):99-117. doi: 10.1089/jir.2010.0097. Epub 2010 Dec 23. J Interferon Cytokine Res. 2011. PMID: 21182352 Free PMC article. Review. - Editing of Cellular Self-RNAs by Adenosine Deaminase ADAR1 Suppresses Innate Immune Stress Responses.
George CX, Ramaswami G, Li JB, Samuel CE. George CX, et al. J Biol Chem. 2016 Mar 18;291(12):6158-68. doi: 10.1074/jbc.M115.709014. Epub 2016 Jan 27. J Biol Chem. 2016. PMID: 26817845 Free PMC article. - Adenosine Deaminases Acting on RNA (ADARs) and Viral Infections.
Pfaller CK, George CX, Samuel CE. Pfaller CK, et al. Annu Rev Virol. 2021 Sep 29;8(1):239-264. doi: 10.1146/annurev-virology-091919-065320. Epub 2021 Apr 21. Annu Rev Virol. 2021. PMID: 33882257 - Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral.
Samuel CE. Samuel CE. Virology. 2011 Mar 15;411(2):180-93. doi: 10.1016/j.virol.2010.12.004. Epub 2011 Jan 5. Virology. 2011. PMID: 21211811 Free PMC article. Review.
Cited by
- _N_6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling.
Kim GW, Imam H, Khan M, Siddiqui A. Kim GW, et al. J Biol Chem. 2020 Sep 11;295(37):13123-13133. doi: 10.1074/jbc.RA120.014260. Epub 2020 Jul 27. J Biol Chem. 2020. PMID: 32719095 Free PMC article. - Next-Generation Sequencing for Confronting Virus Pandemics.
Quer J, Colomer-Castell S, Campos C, Andrés C, Piñana M, Cortese MF, González-Sánchez A, Garcia-Cehic D, Ibáñez M, Pumarola T, Rodríguez-Frías F, Antón A, Tabernero D. Quer J, et al. Viruses. 2022 Mar 14;14(3):600. doi: 10.3390/v14030600. Viruses. 2022. PMID: 35337007 Free PMC article. Review. - Identification of the RNA Pseudoknot within the 3' End of the Porcine Reproductive and Respiratory Syndrome Virus Genome as a Pathogen-Associated Molecular Pattern To Activate Antiviral Signaling via RIG-I and Toll-Like Receptor 3.
Xie S, Chen XX, Qiao S, Li R, Sun Y, Xia S, Wang LJ, Luo X, Deng R, Zhou EM, Zhang GP. Xie S, et al. J Virol. 2018 May 29;92(12):e00097-18. doi: 10.1128/JVI.00097-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618647 Free PMC article. - Mutational Pressure in Zika Virus: Local ADAR-Editing Areas Associated with Pauses in Translation and Replication.
Khrustalev VV, Khrustaleva TA, Sharma N, Giri R. Khrustalev VV, et al. Front Cell Infect Microbiol. 2017 Feb 22;7:44. doi: 10.3389/fcimb.2017.00044. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28275585 Free PMC article. - Sensing of transposable elements by the antiviral innate immune system.
Gazquez-Gutierrez A, Witteveldt J, R Heras S, Macias S. Gazquez-Gutierrez A, et al. RNA. 2021 Apr 22;27(7):735-52. doi: 10.1261/rna.078721.121. Online ahead of print. RNA. 2021. PMID: 33888553 Free PMC article.
References
- Bass BL, Weintraub H. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55(6):1089–1098 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials