Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain - PubMed (original) (raw)
Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain
Corey M Hudson et al. PLoS One. 2014.
Abstract
Multidrug-resistant Enterobacteriaceae are emerging as a serious infectious disease challenge. These strains can accumulate many antibiotic resistance genes though horizontal transfer of genetic elements, those for β-lactamases being of particular concern. Some β-lactamases are active on a broad spectrum of β-lactams including the last-resort carbapenems. The gene for the broad-spectrum and carbapenem-active metallo-β-lactamase NDM-1 is rapidly spreading. We present the complete genome of Klebsiella pneumoniae ATCC BAA-2146, the first U.S. isolate found to encode NDM-1, and describe its repertoire of antibiotic-resistance genes and mutations, including genes for eight β-lactamases and 15 additional antibiotic-resistance enzymes. To elucidate the evolution of this rich repertoire, the mobile elements of the genome were characterized, including four plasmids with varying degrees of conservation and mosaicism and eleven chromosomal genomic islands. One island was identified by a novel phylogenomic approach, that further indicated the cps-lps polysaccharide synthesis locus, where operon translocation and fusion was noted. Unique plasmid segments and mosaic junctions were identified. Plasmid-borne blaCTX-M-15 was transposed recently to the chromosome by ISEcp1. None of the eleven full copies of IS26, the most frequent IS element in the genome, had the expected 8-bp direct repeat of the integration target sequence, suggesting that each copy underwent homologous recombination subsequent to its last transposition event. Comparative analysis likewise indicates IS26 as a frequent recombinational junction between plasmid ancestors, and also indicates a resolvase site. In one novel use of high-throughput sequencing, homologously recombinant subpopulations of the bacterial culture were detected. In a second novel use, circular transposition intermediates were detected for the novel insertion sequence ISKpn21 of the ISNCY family, suggesting that it uses the two-step transposition mechanism of IS3. Robust genome-based phylogeny showed that a unified Klebsiella cluster contains Enterobacter aerogenes and Raoultella, suggesting the latter genus should be abandoned.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Klebsiella phylogeny.
Tree for 108 genomes based on a 2.93-Mbp alignment, rooted at the midpoint of the outgroup (Ecl/Yre) branch. Nodes with <30% bootstrap support were combined forming the multifurcated dashed line; otherwise support values are shown only when <100%. Brackets: Kpn multilocus sequence type (ST). Inset: enlargement of the “core Kpn” phylogeny. Kpn2146 falls in a clade containing fellow ST11 strains Kpn JM45 and Kpn HS11286 and a tight clade (circled) of ST258 and ST512 strains. The ST258/ST512 clade is heavily sequenced, and represented here with only five of its most diverse members. Bold: complete genomes used for phyloblocks analysis. Species name abbreviations: Kpn, K. pneumoniae; Ksp, K. sp.; Kpl, K. cf. planticola; Kox, K. oxytoca; Kva, K. variicola; Eae, Enterobacter aerogenes; Ecl, E. cloacae; Ror, Raoultella ornithinolytica; Yre, Yokanella regensburgei.
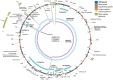
Figure 2. pNDM-US.
Key, color coding of genes, mobile elements, and unique regions and juxtapositions, with additional colors for non-gene features. Inner ring, representative long matches to other plasmids. abR, antibiotic resistance.
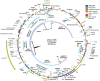
Figure 3. pKpn2146c.
Key, color coding of genes, mobile elements, and unique regions and juxtapositions, with additional colors for non-gene features. Inner ring, representative long matches to other plasmids. Innermost black arrows, recent recombination events. HR, homologous recombination; abR, antibiotic resistance.
Figure 4. pKpn2146b.
Key, color coding of genes, mobile elements, and unique regions and juxtapositions, with additional colors for non-gene features. Inner ring, representative long matches to other plasmids. Innermost black arrows, recent recombination events. HR, homologous recombination; abR, antibiotic resistance.
Figure 5. Learned phyloblocks identify a new island and the highly variable capsular polysaccharide and lipopolysaccharide synthesis gene cluster (cps-lps).
Nonubiquity phyloblocks: those missing in at least one of the 11 reference chromosomes. Complex phyloblocks: those requiring more than one gain/loss event to reconcile the phylotype with the genome tree of Fig. 1. As a percentage of their combined 411 kbp, the learned phyloblocks mapped either to the training islands (81.9%), the two newly indicated regions (12.0%), insertion sequences (2.1%), or to small scattered regions that did not show hallmarks of islands (4.0%). Red segments: the 11 final islands (including a tandem array of Kpn21L and Kpn11L). Circles, the two newly indicated regions.
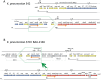
Figure 6. Operon translocation and fusion at the _cps_-lps polysaccharide synthesis locus.
The cps P1, P2 and P3 promoters are taken from , while a promoter (P_lps_) has been mapped in K. pneumoniae MGH 78578 to the intergenic space between uge and the first lps gene . A) The cps-lps region of K. pneumoniae 342, which is typical of Klebsiella. Genes of cps are in yellow (common in most strains) or blue (varying in gene identity, count, and order); genes of lps are in red. The manCB unit (orange arrows) is occasionally found in cps, and occasionally in lps, and here unusually in both. The diamond represents the JUMPstart DNA/RNA motif at whose ops sequence RfaH is loaded onto the elongating RNA polymerase in place of NusG, preventing Rho-based termination for the small number of long transcription units that are controlled by _ops_-RfaH, and physically coupling the elongating RNA polymerase to the trailing ribosome . B) Kpn2146 cps-lps. The boxed cps P3 unit has been deleted from its usual site, and moreover translocated to a nearby position, apparently by transposition and/or homologous recombination mechanisms; note the complex pattern of surrounding IS insertions and the directly repeated flanking sequence copies (gray arrows).ΔIS, incomplete IS copy; dotted lines, gene or IS interrupted by ISs; GT, glucosyl transferase, Hyp, hypothetical.
Similar articles
- Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both _bla_KPC and _bla_CTX-M Integrated in the Chromosome.
Huang W, Wang G, Sebra R, Zhuge J, Yin C, Aguero-Rosenfeld ME, Schuetz AN, Dimitrova N, Fallon JT. Huang W, et al. Antimicrob Agents Chemother. 2017 Jun 27;61(7):e00076-17. doi: 10.1128/AAC.00076-17. Print 2017 Jul. Antimicrob Agents Chemother. 2017. PMID: 28438939 Free PMC article. - Genomic Characterization of Carbapenemase-Producing Klebsiella pneumoniae with Chromosomally Carried _bla_NDM-1.
Sakamoto N, Akeda Y, Sugawara Y, Takeuchi D, Motooka D, Yamamoto N, Laolerd W, Santanirand P, Hamada S. Sakamoto N, et al. Antimicrob Agents Chemother. 2018 Nov 26;62(12):e01520-18. doi: 10.1128/AAC.01520-18. Print 2018 Dec. Antimicrob Agents Chemother. 2018. PMID: 30323033 Free PMC article. - Multiple β-Lactam Resistance Gene-Carrying Plasmid Harbored by Klebsiella quasipneumoniae Isolated from Urban Sewage in Japan.
Suzuki Y, Ida M, Kubota H, Ariyoshi T, Murakami K, Kobayashi M, Kato R, Hirai A, Suzuki J, Sadamasu K. Suzuki Y, et al. mSphere. 2019 Sep 25;4(5):e00391-19. doi: 10.1128/mSphere.00391-19. mSphere. 2019. PMID: 31554719 Free PMC article. - Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance.
Pitout JD, Nordmann P, Poirel L. Pitout JD, et al. Antimicrob Agents Chemother. 2015 Oct;59(10):5873-84. doi: 10.1128/AAC.01019-15. Epub 2015 Jul 13. Antimicrob Agents Chemother. 2015. PMID: 26169401 Free PMC article. Review. - IS_26_ and the IS_26_ family: versatile resistance gene movers and genome reorganizers.
Harmer CJ, Hall RM. Harmer CJ, et al. Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0011922. doi: 10.1128/mmbr.00119-22. Epub 2024 Mar 4. Microbiol Mol Biol Rev. 2024. PMID: 38436262 Review.
Cited by
- Antibiotic use and emerging resistance: how can resource-limited countries turn the tide?
Bebell LM, Muiru AN. Bebell LM, et al. Glob Heart. 2014 Sep;9(3):347-58. doi: 10.1016/j.gheart.2014.08.009. Epub 2014 Oct 31. Glob Heart. 2014. PMID: 25667187 Free PMC article. Review. - Detection of a New Resistance-Mediating Plasmid Chimera in a _bla_OXA-48-Positive Klebsiella pneumoniae Strain at a German University Hospital.
Schwanbeck J, Bohne W, Hasdemir U, Groß U, Pfeifer Y, Bunk B, Riedel T, Spröer C, Overmann J, Frickmann H, Zautner AE. Schwanbeck J, et al. Microorganisms. 2021 Mar 31;9(4):720. doi: 10.3390/microorganisms9040720. Microorganisms. 2021. PMID: 33807212 Free PMC article. - First report of coexistence of blaKPC-2 and blaNDM-1 in carbapenem-resistant clinical isolates of Klebsiella aerogenes in Brazil.
Rodrigues SH, Nunes GD, Soares GG, Ferreira RL, Damas MSF, Laprega PM, Shilling RE, Campos LC, da Costa AS, Malavazi I, da Cunha AF, Pranchevicius MDS. Rodrigues SH, et al. Front Microbiol. 2024 Feb 14;15:1352851. doi: 10.3389/fmicb.2024.1352851. eCollection 2024. Front Microbiol. 2024. PMID: 38426065 Free PMC article. - Preharvest Transmission Routes of Fresh Produce Associated Bacterial Pathogens with Outbreak Potentials: A Review.
Iwu CD, Okoh AI. Iwu CD, et al. Int J Environ Res Public Health. 2019 Nov 11;16(22):4407. doi: 10.3390/ijerph16224407. Int J Environ Res Public Health. 2019. PMID: 31717976 Free PMC article. Review. - Small Klebsiella pneumoniae Plasmids: Neglected Contributors to Antibiotic Resistance.
Ramirez MS, Iriarte A, Reyes-Lamothe R, Sherratt DJ, Tolmasky ME. Ramirez MS, et al. Front Microbiol. 2019 Sep 20;10:2182. doi: 10.3389/fmicb.2019.02182. eCollection 2019. Front Microbiol. 2019. PMID: 31616398 Free PMC article.
References
- Gupta N, Limbago BM, Patel JB, Kallen AJ (2011) Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53: 60–67. - PubMed
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. (2009) Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53: 5046–5054. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was funded by Sandia National Laboratories' Grand Challenge LDRD (Laboratory-Directed Research and Development, grant number 165683) program. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the U.S. Department of Energy's National Nuclear Security Administration under Contract DE-AC04-94AL85000. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases