Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome - PubMed (original) (raw)
. 2014 Sep;24(9):1517-25.
doi: 10.1101/gr.168245.113. Epub 2014 Jun 6.
Christina D Moon 2, Sinead C Leahy 2, Dongwan Kang 1, Jeff Froula 1, Sandra Kittelmann 2, Christina Fan 1, Samuel Deutsch 1, Dragana Gagic 2, Henning Seedorf 2, William J Kelly 2, Renee Atua 2, Carrie Sang 2, Priya Soni 2, Dong Li 2, Cesar S Pinares-Patiño 2, John C McEwan 2, Peter H Janssen 2, Feng Chen 1, Axel Visel 3, Zhong Wang 3, Graeme T Attwood 2, Edward M Rubin 4
Affiliations
- PMID: 24907284
- PMCID: PMC4158751
- DOI: 10.1101/gr.168245.113
Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome
Weibing Shi et al. Genome Res. 2014 Sep.
Abstract
Ruminant livestock represent the single largest anthropogenic source of the potent greenhouse gas methane, which is generated by methanogenic archaea residing in ruminant digestive tracts. While differences between individual animals of the same breed in the amount of methane produced have been observed, the basis for this variation remains to be elucidated. To explore the mechanistic basis of this methane production, we measured methane yields from 22 sheep, which revealed that methane yields are a reproducible, quantitative trait. Deep metagenomic and metatranscriptomic sequencing demonstrated a similar abundance of methanogens and methanogenesis pathway genes in high and low methane emitters. However, transcription of methanogenesis pathway genes was substantially increased in sheep with high methane yields. These results identify a discrete set of rumen methanogens whose methanogenesis pathway transcription profiles correlate with methane yields and provide new targets for CH4 mitigation at the levels of microbiota composition and transcriptional regulation.
© 2014 Shi et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
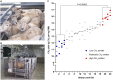
Figure 1.
The measurement of CH4 yields in sheep. (A) New Zealand sheep used for this study. (B) CH4 yields from the sheep in grams of CH4/kg dry matter intake (DMI) were measured using open-circuit respiration chambers (
http://www.globalresearchalliance.org
). (C) CH4 yield measurements from 22 sheep (each with two time points) sorted by mean values. Four high (red) and four low (blue) emitters are selected for further study. _P_-value indicates the statistical significance of the differences in CH4 yield between the two selected groups.
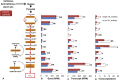
Figure 2.
Comparison of relative abundance of different microbial populations in low and high CH4 yield sheep. (A) Relative abundance of microbial domains in low and high CH4 yield sheep. (B) Relative abundance of methanogenic and nonmethanogenic archaea in low and high CH4 yield sheep. (C) Relative abundance of classes of CH4-producing Euryarchaeota in low and high CH4 yield sheep. (NS) No statistical difference in Wilcoxon rank-sum test in each subgroup.
Figure 3.
Comparisons of gene and transcript abundance for enzymes involved in methanogenesis between high and low CH4 yield sheep. (A) Diagram of CO2/H2 methanogenesis pathway shows enzymes involved in each biochemical reaction. (B,C) Gene (B) and transcript (C) abundance for each enzyme. (D) Transcriptions per gene for each enzyme. (RPM) Reads per million; (NS) no statistical significance in Wilcoxon rank-sum test; (*) P < 0.05; (**) P < 0.01. Error bars, SE.
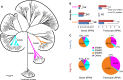
Figure 4.
Phylogenetic analysis of methanogens in sheep rumen. (A) A phylogenetic tree constructed based on full-length methyl coenzyme M reductase alpha subunit (McrA/MrtA) protein sequences. Known McrA/MrtA proteins from NCBI are shown in black; new ones from this study, in color. (B) Genes and transcripts for three groups of identified sheep rumen methanogens. (RPM) Reads per million; (NS) no statistical significance in Wilcoxon rank-sum test; (*) P < 0.05; (**) P < 0.01. Error bars, SE. (C) Relative contribution of each group of sheep rumen methanogens to the overall abundance (RPM) of genes and transcripts in low and high CH4 yield sheep. The sizes of each pie indicate the abundance of genes/transcripts.
Similar articles
- The rumen microbial metagenome associated with high methane production in cattle.
Wallace RJ, Rooke JA, McKain N, Duthie CA, Hyslop JJ, Ross DW, Waterhouse A, Watson M, Roehe R. Wallace RJ, et al. BMC Genomics. 2015 Oct 23;16:839. doi: 10.1186/s12864-015-2032-0. BMC Genomics. 2015. PMID: 26494241 Free PMC article. - Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation.
Kamke J, Kittelmann S, Soni P, Li Y, Tavendale M, Ganesh S, Janssen PH, Shi W, Froula J, Rubin EM, Attwood GT. Kamke J, et al. Microbiome. 2016 Oct 19;4(1):56. doi: 10.1186/s40168-016-0201-2. Microbiome. 2016. PMID: 27760570 Free PMC article. - Gene and transcript abundances of bacterial type III secretion systems from the rumen microbiome are correlated with methane yield in sheep.
Kamke J, Soni P, Li Y, Ganesh S, Kelly WJ, Leahy SC, Shi W, Froula J, Rubin EM, Attwood GT. Kamke J, et al. BMC Res Notes. 2017 Aug 8;10(1):367. doi: 10.1186/s13104-017-2671-0. BMC Res Notes. 2017. PMID: 28789673 Free PMC article. - Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies.
Leahy SC, Kelly WJ, Ronimus RS, Wedlock N, Altermann E, Attwood GT. Leahy SC, et al. Animal. 2013 Jun;7 Suppl 2:235-43. doi: 10.1017/S1751731113000700. Animal. 2013. PMID: 23739466 Review. - Symposium review: Understanding the role of the rumen microbiome in enteric methane mitigation and productivity in dairy cows.
Pitta D, Indugu N, Narayan K, Hennessy M. Pitta D, et al. J Dairy Sci. 2022 Oct;105(10):8569-8585. doi: 10.3168/jds.2021-21466. Epub 2022 Mar 26. J Dairy Sci. 2022. PMID: 35346473 Review.
Cited by
- A metagenomic catalogue of the ruminant gut archaeome.
Mi J, Jing X, Ma C, Shi F, Cao Z, Yang X, Yang Y, Kakade A, Wang W, Long R. Mi J, et al. Nat Commun. 2024 Nov 7;15(1):9609. doi: 10.1038/s41467-024-54025-3. Nat Commun. 2024. PMID: 39505912 Free PMC article. - Unique rumen micromorphology and microbiota-metabolite interactions: features and strategies for Tibetan sheep adaptation to the plateau.
Chen Q, Sha Y, Liu X, He Y, Chen X, Yang W, Gao M, Huang W, Wang J, He J, Wang L. Chen Q, et al. Front Microbiol. 2024 Oct 9;15:1471732. doi: 10.3389/fmicb.2024.1471732. eCollection 2024. Front Microbiol. 2024. PMID: 39444691 Free PMC article. - Integrating genome- and transcriptome-wide association studies to uncover the host-microbiome interactions in bovine rumen methanogenesis.
Wang W, Wei Z, Li Z, Ren J, Song Y, Xu J, Liu A, Li X, Li M, Fan H, Jin L, Niyazbekova Z, Wang W, Gao Y, Jiang Y, Yao J, Li F, Wu S, Wang Y. Wang W, et al. Imeta. 2024 Sep 3;3(5):e234. doi: 10.1002/imt2.234. eCollection 2024 Oct. Imeta. 2024. PMID: 39429883 Free PMC article. - Comprehensive profile of the companion animal gut microbiome integrating reference-based and reference-free methods.
Branck T, Hu Z, Nickols WA, Walsh AM, Bhosle A, Short MI, Nearing JT, Asnicar F, McIver LJ, Maharjan S, Rahnavard A, Louyakis AS, Badri DV, Brockel C, Thompson KN, Huttenhower C. Branck T, et al. ISME J. 2024 Jan 8;18(1):wrae201. doi: 10.1093/ismejo/wrae201. ISME J. 2024. PMID: 39394961 Free PMC article. - Tea Polyphenols Inhibit Methanogenesis and Improve Rumen Epithelial Transport in Dairy Cows.
Teng Z, Liu S, Zhang L, Zhang L, Liu S, Fu T, Zhang N, Gao T. Teng Z, et al. Animals (Basel). 2024 Sep 4;14(17):2569. doi: 10.3390/ani14172569. Animals (Basel). 2024. PMID: 39272354 Free PMC article.
References
- Benchaar C, Pomar C, Chiquette J. 2001. Evaluation of dietary strategies to reduce methane production in ruminants: a modelling approach. Can J Anim Sci 81: 563–574
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300
- Boone DR, Whitman WB, Rouviere P. 1993. Diversity and taxonomy of methanogens. In Methanogenesis: ecology, physiology, biochemistry and genetics (ed. Ferry JG), pp. 35–80. Chapman and Hall, London, UK.
- Buddle BM, Denis M, Attwood GT, Altermann E, Janssen PH, Ronimus RS, Pinares-Patino CS, Muetzel S, Wedlock DN. 2011. Strategies to reduce methane emissions from farmed ruminants grazing on pasture. Vet J 188: 11–17 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases