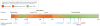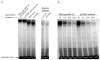Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population - PubMed (original) (raw)
. 2014 Jun 11;311(22):2305-14.
doi: 10.1001/jama.2014.6511.
Karol Estrada 1, Ingvild Aukrust 2, Lise Bjørkhaug 3, Noël P Burtt 4, Josep M Mercader 5, Humberto García-Ortiz 6, Alicia Huerta-Chagoya 7, Hortensia Moreno-Macías 8, Geoffrey Walford 9, Jason Flannick 10, Amy L Williams 11, María J Gómez-Vázquez 12, Juan C Fernandez-Lopez 6, Angélica Martínez-Hernández 6, Silvia Jiménez-Morales 6, Federico Centeno-Cruz 6, Elvia Mendoza-Caamal 6, Cristina Revilla-Monsalve 13, Sergio Islas-Andrade 13, Emilio J Córdova 6, Xavier Soberón 6, María E González-Villalpando 14, E Henderson 15, Lynne R Wilkens 16, Loic Le Marchand 16, Olimpia Arellano-Campos 12, Maria L Ordóñez-Sánchez 12, Maribel Rodríguez-Torres 12, Rosario Rodríguez-Guillén 12, Laura Riba 7, Laeya A Najmi 17, Suzanne B R Jacobs 4, Timothy Fennell 18, Stacey Gabriel 18, Pierre Fontanillas 4, Craig L Hanis 19, Donna M Lehman 20, Christopher P Jenkinson 20, Hanna E Abboud 20, Graeme I Bell 21, Maria L Cortes 22, Michael Boehnke 23, Clicerio González-Villalpando 14, Lorena Orozco 6, Christopher A Haiman 15, Teresa Tusié-Luna 24, Carlos A Aguilar-Salinas 12, David Altshuler 25, Pål R Njølstad 3, Jose C Florez 25, Daniel G MacArthur 26
Collaborators, Affiliations
- PMID: 24915262
- PMCID: PMC4425850
- DOI: 10.1001/jama.2014.6511
Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population
SIGMA Type 2 Diabetes Consortium et al. JAMA. 2014.
Erratum in
- JAMA. 2014 Nov 12:312(18):1932. Jiménez-Morales, Silvia[Added]
Abstract
Importance: Latino populations have one of the highest prevalences of type 2 diabetes worldwide.
Objectives: To investigate the association between rare protein-coding genetic variants and prevalence of type 2 diabetes in a large Latino population and to explore potential molecular and physiological mechanisms for the observed relationships.
Design, setting, and participants: Whole-exome sequencing was performed on DNA samples from 3756 Mexican and US Latino individuals (1794 with type 2 diabetes and 1962 without diabetes) recruited from 1993 to 2013. One variant was further tested for allele frequency and association with type 2 diabetes in large multiethnic data sets of 14,276 participants and characterized in experimental assays.
Main outcome and measures: Prevalence of type 2 diabetes. Secondary outcomes included age of onset, body mass index, and effect on protein function.
Results: A single rare missense variant (c.1522G>A [p.E508K]) was associated with type 2 diabetes prevalence (odds ratio [OR], 5.48; 95% CI, 2.83-10.61; P = 4.4 × 10(-7)) in hepatocyte nuclear factor 1-α (HNF1A), the gene responsible for maturity onset diabetes of the young type 3 (MODY3). This variant was observed in 0.36% of participants without type 2 diabetes and 2.1% of participants with it. In multiethnic replication data sets, the p.E508K variant was seen only in Latino patients (n = 1443 with type 2 diabetes and 1673 without it) and was associated with type 2 diabetes (OR, 4.16; 95% CI, 1.75-9.92; P = .0013). In experimental assays, HNF-1A protein encoding the p.E508K mutant demonstrated reduced transactivation activity of its target promoter compared with a wild-type protein. In our data, carriers and noncarriers of the p.E508K mutation with type 2 diabetes had no significant differences in compared clinical characteristics, including age at onset. The mean (SD) age for carriers was 45.3 years (11.2) vs 47.5 years (11.5) for noncarriers (P = .49) and the mean (SD) BMI for carriers was 28.2 (5.5) vs 29.3 (5.3) for noncarriers (P = .19).
Conclusions and relevance: Using whole-exome sequencing, we identified a single low-frequency variant in the MODY3-causing gene HNF1A that is associated with type 2 diabetes in Latino populations and may affect protein function. This finding may have implications for screening and therapeutic modification in this population, but additional studies are required.
Figures
Figure 1. Discovery and Replication of the HNF1A p.E508K Variant
Forest plot showing odds ratio estimates and 95% confidence intervals at p.E508K (squared boxes) from the 4 SIGMA studies, the SIGMA pooled mega-analysis, the replication studies, and the overall meta-analysis. Odds ratios for the meta-analyses are represented with a diamond. SIGMA mega-analysis represents the combined results from the 4 SIGMA studies. DMS indicates Diabetes in Mexico Study; MCDS, Mexico City Diabetes Study; MEC, Multiethnic Cohort; UIDS, Universidad Nacional Autónoma de México/Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán Diabetes Study; T2D-GENES, Type 2 Diabetes Genetic Exploration by Next-Generation Sequencing in Multi-Ethnic Samples. aRepresents data from the current article.
Figure 2. The HNF-1A Protein With a Heat Map of Diabetes-Associated Mutations
The dimerization, DNA binding, and transactivation domains of the HNF-1A protein– are highlighted. The position of the p.E508K mutation is shown as well as a common variant (p.I27L), MODY3 mutations studied (p.P112L, p.R229Q, p.P379fsdelCT, p.P447L, p.Q466X), and a rare variant associated with type 2 diabetes (p.M490T). The overlaid heat map illustrates how many of the amino acid residues of each HNF-1A domain have been reported to be mutated and hence due to the monogenic diabetes form MODY3. Domain areas in red have a higher concentration of reported mutations than areas in orange and green. Pseudo POU indicates protein domain that includes short sequence motifs similar to regions in the POU family of transcriptional activators; Homeo, protein homeodomain that binds DNA in a sequence-specific manner.
Figure 3. Transcriptional Activation of HNF-1A p.E508K as Measured by the Expression of the Firefly Luciferase Reporter Gene
HeLa cells were transient transfected with nonmutant or mutant HNF1A plasmids and reporter plasmids pGL3-RA and pRL-SV40. Measurements are given in fold activity relative to wild-type. Each point represents the mean (error bars indicate 95% CIs) of 9 readings. TA indicates variants that affect the transactivation domain; DNA bind, the DNA binding domain; and pcDNA3.1, the empty pcDNA3.1 vector. All values were P < .05 compared with wild-type activity.
Figure 4. DNA Binding of HNF-1A p.E508K to the Rat Albumin Promoter as Studied by Electrophoretic Mobility Shift Assay
Xpress-epitope-tagged wild-type and p.E508K mutant proteins incubated with a radiolabeled DNA fragment containing the HNF-1A-binding site in the rat albumin promoter. A, Two HNF-1A mutants (p.P112L and p.R229Q) with impaired DNA-binding were included as negative controls. Addition of the anti-Xpress antibody induced a supershift (a reduction in mobility of protein-DNA complex due to antibody binding, relative to protein-DNA complex alone) for the DNA-protein complexes, confirming the identity of HNF-1A within the complexes. B, A competition assay was performed by adding increasing amounts (0x, 10x, 50x, or 100x) of radiolabeled DNA fragment, confirming the identity of the radiolabeled probe.
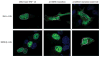
Figure 5. Intracellular Localization of HNF-1A p.E508K in Transiently Transfected HeLa cells and MIN6 β cells
Cells were transfected for 48 hours and Xpress-epitope-tagged HNF-1A proteins detected with anti-Xpress antibody and Alexa488 (green). DNA staining (DAPI) is shown in blue. A previously reported HNF-1A mutant, p.Q466X, with impaired nuclear localization was included as a control. For the purpose of clarity, the nuclei have been marked with a solid white line. To illustrate cytosolic accumulation, the cell membrane has been marked with a dotted white line for mutants p.E508K and p.Q466X.
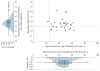
Figure 6. Phenotypic Distribution of p.E508K Carriers
The scatterplot shows the age of onset and the body mass index (BMI) for each p.E508K carrier (filled circle) with type 2 diabetes in the discovery studies with data on age of onset and BMI available (n = 29). The vertical and horizontal lines represent classical thresholds for the clinical diagnosis of MODY3 (age of onset <25 years and BMI<25). Histograms showing distributions of BMI and age of diabetes onset 1274 SIGMA discovery cohort participants (p.E508K carriers and noncarriers with Type 2 diabetes) are shown on the left and below the scatterplot. In the box-and-whisker plots, the central horizontal line indicates median, with box extremes indicating the first and third quartiles. The whiskers indicate maximum and minimum values after removal of outliers (unfilled circles).
Similar articles
- Mexican Carriers of the HNF1A p.E508K Variant Do Not Experience an Enhanced Response to Sulfonylureas.
Martagón AJ, Bello-Chavolla OY, Arellano-Campos O, Almeda-Valdés P, Walford GA, Cruz-Bautista I, Gómez-Velasco DV, Mehta R, Muñoz-Hernández L, Sevilla-González M, Viveros-Ruiz TL, Ordoñez-Sánchez ML, Rodríguez-Guillen R, Florez JC, Tusié-Luna MT, Aguilar-Salinas CA; Slim Initiative in Genomic Medicine for the Americas (SIGMA) Type 2 Diabetes Consortium. Martagón AJ, et al. Diabetes Care. 2018 Aug;41(8):1726-1731. doi: 10.2337/dc18-0384. Epub 2018 May 29. Diabetes Care. 2018. PMID: 29844095 - The HNF1A mutant Ala180Val: Clinical challenges in determining causality of a rare HNF1A variant in familial diabetes.
Sagen JV, Bjørkhaug L, Haukanes BI, Grevle L, Molnes J, Nedrebø BG, Søvik O, Njølstad PR, Johansson S, Molven A. Sagen JV, et al. Diabetes Res Clin Pract. 2017 Nov;133:142-149. doi: 10.1016/j.diabres.2017.08.001. Epub 2017 Sep 1. Diabetes Res Clin Pract. 2017. PMID: 28934671 - Structure-function studies of HNF1A (MODY3) gene mutations in South Indian patients with monogenic diabetes.
Balamurugan K, Bjørkhaug L, Mahajan S, Kanthimathi S, Njølstad PR, Srinivasan N, Mohan V, Radha V. Balamurugan K, et al. Clin Genet. 2016 Dec;90(6):486-495. doi: 10.1111/cge.12757. Epub 2016 Mar 4. Clin Genet. 2016. PMID: 26853433 - Novel insights into genetics and clinics of the HNF1A-MODY.
Valkovicova T, Skopkova M, Stanik J, Gasperikova D. Valkovicova T, et al. Endocr Regul. 2019 Apr 1;53(2):110-134. doi: 10.2478/enr-2019-0013. Endocr Regul. 2019. PMID: 31517624 Review. - Meta-analysis of HNF1A-MODY3 variants among human population.
Behl R, Malhotra N, Joshi V, Poojary S, Middha S, Gupta S, Olaonipekun AB, Okoye I, Wagh B, Biswas D, Aginah C, Saini B, Nwanya C, Ugwu S, Anthony MM, Fang XS, Foluso O, Ibrahim AT. Behl R, et al. J Diabetes Metab Disord. 2022 Feb 2;21(1):1037-1046. doi: 10.1007/s40200-022-00975-8. eCollection 2022 Jun. J Diabetes Metab Disord. 2022. PMID: 35673428 Free PMC article. Review.
Cited by
- Exome Sequence Data of Eight SLC Transporters Reveal That SLC22A1 and SLC22A3 Variants Alter Metformin Pharmacokinetics and Glycemic Control.
Morales-Rivera MI, Alemón-Medina R, Martínez-Hernández A, Contreras-Cubas C, Altamirano-Bustamante NF, Gómez-Garduño J, Mendoza-Caamal EC, Nuñez-González JO, García-Álvarez R, Revilla-Monsalve C, Valcarcel-Gamiño JA, Villafan-Bernal JR, Centeno-Cruz F, García-Ortiz H, Barajas-Olmos F, Orozco L. Morales-Rivera MI, et al. Pharmaceuticals (Basel). 2024 Oct 17;17(10):1385. doi: 10.3390/ph17101385. Pharmaceuticals (Basel). 2024. PMID: 39459024 Free PMC article. - Rare variant analyses in 51,256 type 2 diabetes cases and 370,487 controls reveal the pathogenicity spectrum of monogenic diabetes genes.
Huerta-Chagoya A, Schroeder P, Mandla R, Li J, Morris L, Vora M, Alkanaq A, Nagy D, Szczerbinski L, Madsen JGS, Bonàs-Guarch S, Mollandin F, Cole JB, Porneala B, Westerman K, Li JH, Pollin TI, Florez JC, Gloyn AL, Carey DJ, Cebola I, Mirshahi UL, Manning AK, Leong A, Udler M, Mercader JM. Huerta-Chagoya A, et al. Nat Genet. 2024 Nov;56(11):2370-2379. doi: 10.1038/s41588-024-01947-9. Epub 2024 Oct 8. Nat Genet. 2024. PMID: 39379762 Free PMC article. - Analysis of an adult diabetes mellitus caused by a rare mutation of the gene: A case report.
Li WX, Xu LL, Liu CF, Dong BZ, Wang YY. Li WX, et al. World J Clin Cases. 2024 Jul 6;12(19):3942-3949. doi: 10.12998/wjcc.v12.i19.3942. World J Clin Cases. 2024. PMID: 38994305 Free PMC article. - KRT81 and HNF1A expression in pancreatic ductal adenocarcinoma: investigation of predictive and prognostic value of immunohistochemistry-based subtyping.
Rao J, Sinn M, Pelzer U, Riess H, Oettle H, Demir IE, Friess H, Jäger C, Steiger K, Muckenhuber A. Rao J, et al. J Pathol Clin Res. 2024 May;10(3):e12377. doi: 10.1002/2056-4538.12377. J Pathol Clin Res. 2024. PMID: 38750616 Free PMC article. - CFTR pathogenic variants spectrum in a cohort of Mexican patients with cystic fibrosis.
Martínez-Hernández A, Mendoza-Caamal EC, Mendiola-Vidal NG, Barajas-Olmos F, Villafan-Bernal JR, Jiménez-Ruiz JL, Monge-Cazares T, García-Ortiz H, Cubas CC, Centeno-Cruz F, Alaez-Verson C, Ortega-Torres S, Luna-Castañeda ADC, Baca V, Lezana JL, Orozco L. Martínez-Hernández A, et al. Heliyon. 2024 Mar 29;10(7):e28984. doi: 10.1016/j.heliyon.2024.e28984. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38601560 Free PMC article.
References
- Villalpando S, de la Cruz V, Rojas R, et al. Prevalence and distribution of type 2 diabetes mellitus in Mexican adult population. Salud Publica Mex. 2010;52(suppl 1):S19–S26. - PubMed
- Barquera S, Tovar-Guzmán V, Campos-Nonato I, González-Villalpando C, Rivera-Dommarco J. Geography of diabetes mellitus mortality in Mexico. Arch Med Res. 2003;34(5):407–414. - PubMed
- Cowie CC, Rust KF, Byrd-Holt DD, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health And Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29(6):1263–1268. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- U01 DK085501/DK/NIDDK NIH HHS/United States
- U01DK085501/DK/NIDDK NIH HHS/United States
- UM1 CA164973/CA/NCI NIH HHS/United States
- R01 CA80205/CA/NCI NIH HHS/United States
- U01 DK057295/DK/NIDDK NIH HHS/United States
- R01 CA144034/CA/NCI NIH HHS/United States
- R01 DK047482/DK/NIDDK NIH HHS/United States
- R01 DK033665/DK/NIDDK NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- U01 DK062370/DK/NIDDK NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- P01HL045522/HL/NHLBI NIH HHS/United States
- R01 DK053889/DK/NIDDK NIH HHS/United States
- R35CA53890/CA/NCI NIH HHS/United States
- R01 DK098032/DK/NIDDK NIH HHS/United States
- U01DK085526/DK/NIDDK NIH HHS/United States
- R01DK053889/DK/NIDDK NIH HHS/United States
- R01 DK057295/DK/NIDDK NIH HHS/United States
- CA164973/CA/NCI NIH HHS/United States
- R01 DK042273/DK/NIDDK NIH HHS/United States
- R35 CA053890/CA/NCI NIH HHS/United States
- R01 CA055069/CA/NCI NIH HHS/United States
- CA063464/CA/NCI NIH HHS/United States
- CA054281/CA/NCI NIH HHS/United States
- U01 DK085526/DK/NIDDK NIH HHS/United States
- P01 HL045522/HL/NHLBI NIH HHS/United States
- R01DK047482/DK/NIDDK NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- R01 CA080205/CA/NCI NIH HHS/United States
- R01HL24799/HL/NHLBI NIH HHS/United States
- U01 CA164973/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical