The interaction between amyloid-β peptides and anionic lipid membranes containing cholesterol and melatonin - PubMed (original) (raw)
The interaction between amyloid-β peptides and anionic lipid membranes containing cholesterol and melatonin
Hannah Dies et al. PLoS One. 2014.
Abstract
One of the hallmarks of Alzheimer's disease is the formation of senile plaques, primarily consisting of amyloid-β (Aβ) peptides. Peptide-membrane and peptide-lipid interactions are thought to be crucial in this process. We studied the interaction of Aβ₁₋₄₂ and Aβ₂₅₋₃₅ peptides with anionic lipid membranes made of dimyristoylphosphatidylcholine (DMPC) and dimyristoylphosphoserine (DMPS) using X-ray diffraction. We compare the experimentally determined electron densities in the gel state of the membranes with density calculations from peptide structures reported in the Protein Data Bank in order to determine the position of the peptide in the bilayers. The full length peptide Aβ₁₋₄₂ was found to embed in the hydrocarbon core of the anionic lipid bilayers. Two populations were found for the Aβ₂₅₋₃₅ peptide: (1) membrane-bound states in the hydrophilic head group region of the bilayers, where the peptides align parallel to the membranes, and (2) an embedded state in the bilayer center. Aging plays an important role in the development of Alzheimer's, in particular with respect to changes in cholesterol and melatonin levels in the brain tissue. Immiscible cholesterol plaques were created by addition of 30 mol% cholesterol to the anionic membranes. The Aβ₂₅₋₃₅ peptides were found to strongly interact with the lipid bilayers, displacing further cholesterol molecules into the plaques, effectively lowering the cholesterol concentration in the membranes and increasing the total fraction of cholesterol plaques. Addition of 30 mol% melatonin molecules to the anionic membranes drastically reduced the population of the membrane-embedded Aβ state. These results present experimental evidence for an interaction between Aβ peptides, melatonin and cholesterol in lipid membranes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
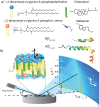
Figure 1. The materials and apparatus used for the experiment.
(a) Schematic representations of DMPC, DMPS, cholesterol, melatonin, amyloid- and amyloid-
and amyloid- molecules. (b) Diagram of the experimental setup used for the X-ray diffraction measurements. Two-dimensional data sets were collected to study molecular structure perpendicular to the solid supported membranes (out-of-plane) and parallel to the membranes (in-plane).
molecules. (b) Diagram of the experimental setup used for the X-ray diffraction measurements. Two-dimensional data sets were collected to study molecular structure perpendicular to the solid supported membranes (out-of-plane) and parallel to the membranes (in-plane).
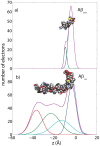
Figure 2. Calculated electron distribution of A and A.
The molecular structures and calculations are based on the PDB structures 1QWP (A ) and 1IYT (A
) and 1IYT (A ). The peptides take helix-kink and helix-kink-helix configurations, respectively, in solution.
). The peptides take helix-kink and helix-kink-helix configurations, respectively, in solution.
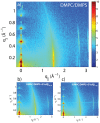
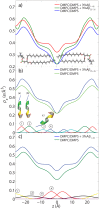
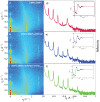
Figure 3. Two-dimensional data for (a) DMPC +3 DMPS membrane, (b) DMPC/DMPS+3 mol% A and (c) DMPC/DMPS+3 mol% A.
The area per lipid is obtained from the position of the lipid acyl chain correlation peak at  1.5 Å−1.
1.5 Å−1.
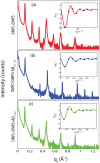
Figure 4. Reflectivity measurements for (a) DMPC/DMPS membrane, (b) DMPC/DMPS+3 mol% A, and (c) DMPC/DMPS+3 mol% A.
Pronounced and equally spaced Bragg peaks were observed, which are indicative to a well ordered lamellar structure. Electron densities were calculated through Fourier transformation of the integrated peak intensities. The insets shows the  function, which was used to assess the phases of the corresponding Fourier components.
function, which was used to assess the phases of the corresponding Fourier components.
Figure 5. Electron density analyses obtained through Fourier transforms of the X-ray reflectivity data.
a) Electron densities for the DMPC/DMPS membrane (green), DMPC/DMPS+3 mol% A (blue), and DMPC/DMPS+3 mol% A
(blue), and DMPC/DMPS+3 mol% A (red). b) The location of the A
(red). b) The location of the A peptide in the DMPC/DMPS membrane, as determined from the difference electron density profile peak positions. c) Gaussian fits to the difference electron density obtained by subtracting the electron density profile of the DMPC/DMPS sample from that of the DMPC/DMPS+3 mol% A
peptide in the DMPC/DMPS membrane, as determined from the difference electron density profile peak positions. c) Gaussian fits to the difference electron density obtained by subtracting the electron density profile of the DMPC/DMPS sample from that of the DMPC/DMPS+3 mol% A sample. A total of 4 peaks in each half of the bilayer are needed to fit the experimental data. The
sample. A total of 4 peaks in each half of the bilayer are needed to fit the experimental data. The  -axis encompasses the full bilayer.
-axis encompasses the full bilayer.
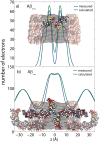
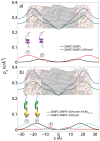
Figure 6. Measured and calculated electron distribution of the membrane-embedded A a) and A b) peptides and their position in the membrane.
Good agreement between calculations and experiments is obtained for a position of A in the hydrocarbon membrane core. The peptide takes a slightly tilted orientation, in agreement with computer simulations . The full length A
in the hydrocarbon membrane core. The peptide takes a slightly tilted orientation, in agreement with computer simulations . The full length A peptide was also found to embed in anionic lipid exclude a membrane-spanning
peptide was also found to embed in anionic lipid exclude a membrane-spanning  -sheet structure, as it was reported from molecular dynamics simulations –.
-sheet structure, as it was reported from molecular dynamics simulations –.
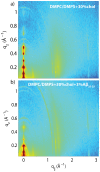
Figure 7. Two-dimensional X-ray scans for the a) DMPC +3 mol% DMPS +30 mol% cholesterol and b) DMPC +3 mol% DMPS 3 mol% A+30 mol% cholesterol membranes.
Data were collected under full hydration of the membranes (100% RH), in their physiologically relevant fluid state.
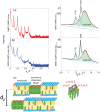
Figure 8. Reflectivity measurements for the a) DMPC/DMPS+30 mol% cholesterol membrane and the b) DMPC/DMPS+3 mol% A+30 mol% cholesterol membrane.
c) and d) show Lorentzian fits to corresponding in-plane data from Figures 7 c and d), scanned under 100 relative humidity. Note that Barrett et al. identified the blue peaks as corresponding to in-plane features of crystalline cholesterol plaques. e) Cartoon of the structure of the multi-lamellar membrane complexes with membranes and coexisting cholesterol plaques and corresponding
relative humidity. Note that Barrett et al. identified the blue peaks as corresponding to in-plane features of crystalline cholesterol plaques. e) Cartoon of the structure of the multi-lamellar membrane complexes with membranes and coexisting cholesterol plaques and corresponding  -spacing. The cholesterol molecules in the plaques form a monoclinic lattice.
-spacing. The cholesterol molecules in the plaques form a monoclinic lattice.
Figure 9. Two-dimensional X-ray data for the a) DMPC/DMPS, b) DMPC/DMPS +30 mol% melatonin, and c) DMPC/DMPS +30 mol% melatonin +3 mol% A samples, with all scans done under full hydration.
Corresponding reflectivities are shown in d), e) and f). The phases can be determined from the  function (shown as insets to the plots) and were used to determine the electron densities plotted in Figure 10.
function (shown as insets to the plots) and were used to determine the electron densities plotted in Figure 10.
Figure 10. Electron densities for the a) DMPC/DMPS, b) DMPC/DMPS +30 mol% melatonin, and c) DMPC/DMPS +30 mol% melatonin +3 mol% A samples.
All samples were scanned under 100% RH. The difference in electron density in a) allows the determination of position of the melatonin molecules in the anionic lipid membranes. The location of the A peptide in the DMPC/DMPS+30 mol% melatonin membrane was determined from the difference electron density profile peak positions in b).
peptide in the DMPC/DMPS+30 mol% melatonin membrane was determined from the difference electron density profile peak positions in b).
Similar articles
- Interaction between Alzheimer's Abeta(25-35) peptide and phospholipid bilayers: the role of cholesterol.
D'Errico G, Vitiello G, Ortona O, Tedeschi A, Ramunno A, D'Ursi AM. D'Errico G, et al. Biochim Biophys Acta. 2008 Dec;1778(12):2710-6. doi: 10.1016/j.bbamem.2008.07.014. Epub 2008 Jul 28. Biochim Biophys Acta. 2008. PMID: 18706389 - The organization of melatonin in lipid membranes.
Dies H, Cheung B, Tang J, Rheinstädter MC. Dies H, et al. Biochim Biophys Acta. 2015 Apr;1848(4):1032-40. doi: 10.1016/j.bbamem.2015.01.006. Epub 2015 Jan 17. Biochim Biophys Acta. 2015. PMID: 25602914 - The Alzheimer's disease Aβ peptide binds to the anionic DMPS lipid bilayer.
Lockhart C, Klimov DK. Lockhart C, et al. Biochim Biophys Acta. 2016 Jun;1858(6):1118-28. doi: 10.1016/j.bbamem.2016.03.001. Epub 2016 Mar 4. Biochim Biophys Acta. 2016. PMID: 26947182 - Understanding Aβ Peptide Binding to Lipid Membranes: A Biophysical Perspective.
Ahyayauch H, Masserini ME, Alonso A, Goñi FM. Ahyayauch H, et al. Int J Mol Sci. 2024 Jun 10;25(12):6401. doi: 10.3390/ijms25126401. Int J Mol Sci. 2024. PMID: 38928107 Free PMC article. Review. - Amyloid beta-protein interactions with membranes and cholesterol: causes or casualties of Alzheimer's disease.
Gibson Wood W, Eckert GP, Igbavboa U, Müller WE. Gibson Wood W, et al. Biochim Biophys Acta. 2003 Mar 10;1610(2):281-90. doi: 10.1016/s0005-2736(03)00025-7. Biochim Biophys Acta. 2003. PMID: 12648781 Review.
Cited by
- Mimicking the Mammalian Plasma Membrane: An Overview of Lipid Membrane Models for Biophysical Studies.
Luchini A, Vitiello G. Luchini A, et al. Biomimetics (Basel). 2020 Dec 31;6(1):3. doi: 10.3390/biomimetics6010003. Biomimetics (Basel). 2020. PMID: 33396534 Free PMC article. Review. - Cholesterol as a key player in amyloid β-mediated toxicity in Alzheimer's disease.
Rudajev V, Novotny J. Rudajev V, et al. Front Mol Neurosci. 2022 Aug 25;15:937056. doi: 10.3389/fnmol.2022.937056. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36090253 Free PMC article. Review. - Cellular absorption of small molecules: free energy landscapes of melatonin binding at phospholipid membranes.
Lu H, Marti J. Lu H, et al. Sci Rep. 2020 Jun 8;10(1):9235. doi: 10.1038/s41598-020-65753-z. Sci Rep. 2020. PMID: 32513935 Free PMC article. - Neutron Scattering at the Intersection of Heart Health Science and Biophysics.
Marquardt D, Alsop RJ, Rheinstädter MC, Harroun TA. Marquardt D, et al. J Cardiovasc Dev Dis. 2015 Jun 2;2(2):125-140. doi: 10.3390/jcdd2020125. J Cardiovasc Dev Dis. 2015. PMID: 29371515 Free PMC article. Review. - Alzheimer's Disease as a Membrane Disorder: Spatial Cross-Talk Among Beta-Amyloid Peptides, Nicotinic Acetylcholine Receptors and Lipid Rafts.
Fabiani C, Antollini SS. Fabiani C, et al. Front Cell Neurosci. 2019 Jul 18;13:309. doi: 10.3389/fncel.2019.00309. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31379503 Free PMC article. Review.
References
- Simons M, Keller P, Dichgans J, Schulz JB (2001) Cholesterol and alzheimers disease is there a link? Neurology 57: 1089–1093. - PubMed
- Casserly I, Topol EJ (2004) Convergence of atherosclerosis and alzheimer's disease: inflammation, cholesterol, and misfolded proteins. The Lancet 363: 1139–1146. - PubMed
- Shobab LA, Hsiung GYR, Feldman HH (2005) Cholesterol in alzheimer's disease. The Lancet Neurology 4: 841–852. - PubMed
- Pappolla MA, Chyan YJ, Poeggeler B, Frangione B, Wilson G, et al. (2000) An assessment of the antioxidant and the antiamyloidogenic properties of melatonin: implications for alzheimer's disease. Journal of Neural Transmission 107: 203–231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical