Epigenetic regulation of bud dormancy events in perennial plants - PubMed (original) (raw)
Review
Epigenetic regulation of bud dormancy events in perennial plants
Gabino Ríos et al. Front Plant Sci. 2014.
Abstract
Release of bud dormancy in perennial plants resembles vernalization in Arabidopsis thaliana and cereals. In both cases, a certain period of chilling is required for accomplishing the reproductive phase, and several transcription factors with the MADS-box domain perform a central regulatory role in these processes. The expression of DORMANCY-ASSOCIATED MADS-box (DAM)-related genes has been found to be up-regulated in dormant buds of numerous plant species, such as poplar, raspberry, leafy spurge, blackcurrant, Japanese apricot, and peach. Moreover, functional evidence suggests the involvement of DAM genes in the regulation of seasonal dormancy in peach. Recent findings highlight the presence of genome-wide epigenetic modifications related to dormancy events, and more specifically the epigenetic regulation of DAM-related genes in a similar way to FLOWERING LOCUS C, a key integrator of vernalization effectors on flowering initiation in Arabidopsis. We revise the most relevant molecular and genomic contributions in the field of bud dormancy, and discuss the increasing evidence for chromatin modification involvement in the epigenetic regulation of seasonal dormancy cycles in perennial plants.
Keywords: DAM gene; bud dormancy; chilling; chromatin; histone modifications.
Figures
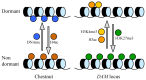
FIGURE 1
General and specific modifications of chromatin in dormant and dormancy-released buds. The following chromatin modifications have been identified in chestnut at the genome level (left), and specifically in the DAM locus in leafy spurge and peach (right): DNA methylation (DNAme), acetylation of histone H4 (H4ac), acetylation of H3 (H3ac), trimethylation of H3 at K4 (H3K4me3), and trimethylation of H3 at K27 (H3K27me3).
Similar articles
- Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner.
Leida C, Conesa A, Llácer G, Badenes ML, Ríos G. Leida C, et al. New Phytol. 2012 Jan;193(1):67-80. doi: 10.1111/j.1469-8137.2011.03863.x. Epub 2011 Sep 7. New Phytol. 2012. PMID: 21899556 - H3K4me3 plays a key role in establishing permissive chromatin states during bud dormancy and bud break in apple.
Chen W, Tamada Y, Yamane H, Matsushita M, Osako Y, Gao-Takai M, Luo Z, Tao R. Chen W, et al. Plant J. 2022 Aug;111(4):1015-1031. doi: 10.1111/tpj.15868. Epub 2022 Jul 1. Plant J. 2022. PMID: 35699670 - I Want to (Bud) Break Free: The Potential Role of DAM and _SVP_-Like Genes in Regulating Dormancy Cycle in Temperate Fruit Trees.
Falavigna VDS, Guitton B, Costes E, Andrés F. Falavigna VDS, et al. Front Plant Sci. 2019 Jan 10;9:1990. doi: 10.3389/fpls.2018.01990. eCollection 2018. Front Plant Sci. 2019. PMID: 30687377 Free PMC article. Review. - Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge.
Horvath DP, Sung S, Kim D, Chao W, Anderson J. Horvath DP, et al. Plant Mol Biol. 2010 May;73(1-2):169-79. doi: 10.1007/s11103-009-9596-5. Epub 2010 Jan 13. Plant Mol Biol. 2010. PMID: 20066557 - Beyond floral initiation: the role of flower bud dormancy in flowering time control of annual plants.
Penfield S. Penfield S. J Exp Bot. 2024 Oct 16;75(19):6056-6062. doi: 10.1093/jxb/erae223. J Exp Bot. 2024. PMID: 38795335 Free PMC article. Review.
Cited by
- Altered expression of a raspberry homologue of VRN1 is associated with disruption of dormancy induction and misregulation of subsets of dormancy-associated genes.
Mateos B, Preedy K, Milne L, Morris J, Hedley PE, Simpson C, Hancock RD, Graham J. Mateos B, et al. J Exp Bot. 2024 Oct 16;75(19):6167-6181. doi: 10.1093/jxb/erae371. J Exp Bot. 2024. PMID: 39243357 Free PMC article. - Characterization of the MIKCC-type MADS-box gene family in blueberry and its possible mechanism for regulating flowering in response to the chilling requirement.
Zhang SL, Wu Y, Zhang XH, Feng X, Wu HL, Zhou BJ, Zhang YQ, Cao M, Hou ZX. Zhang SL, et al. Planta. 2024 Feb 29;259(4):77. doi: 10.1007/s00425-024-04349-7. Planta. 2024. PMID: 38421445 - Molecular Mechanisms of Seasonal Gene Expression in Trees.
Chu X, Wang M, Fan Z, Li J, Yin H. Chu X, et al. Int J Mol Sci. 2024 Jan 30;25(3):1666. doi: 10.3390/ijms25031666. Int J Mol Sci. 2024. PMID: 38338945 Free PMC article. Review. - Uncovering the complex regulatory network of spring bud sprouting in tea plants: insights from metabolic, hormonal, and oxidative stress pathways.
Tang J, Chen Y, Huang C, Li C, Feng Y, Wang H, Ding C, Li N, Wang L, Zeng J, Yang Y, Hao X, Wang X. Tang J, et al. Front Plant Sci. 2023 Oct 23;14:1263606. doi: 10.3389/fpls.2023.1263606. eCollection 2023. Front Plant Sci. 2023. PMID: 37936941 Free PMC article. - Single-Bud Expression Analysis of Bud Dormancy Factors in Peach.
Puertes A, Polat H, Ramón-Núñez LA, González M, Ancillo G, Zuriaga E, Ríos G. Puertes A, et al. Plants (Basel). 2023 Jul 10;12(14):2601. doi: 10.3390/plants12142601. Plants (Basel). 2023. PMID: 37514216 Free PMC article.
References
- Allona I., Ramos A., Ibáñez C., Contreras A., Casado R., Aragoncillo C. (2008). Molecular control of winter dormancy establishment in trees. Span. J. Agric. Res. 6 201–210 10.5424/sjar/200806S1-389 - DOI
- Arend M., Schnitzler J. P., Ehlting B., Hänsch R., Lange T., Rennenberg H., et al. (2009). Expression of the Arabidopsis mutant abi1 gene alters abscisic acid sensitivity, stomatal development, and growth morphology in gray poplars. Plant Physiol. 151 2110–2119 10.1104/pp.109.144956 - DOI - PMC - PubMed
- Arora R., Rowland L. J., Tanino K. (2003). Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 38 911–921
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources