Prediction of protein S-nitrosylation sites based on adapted normal distribution bi-profile Bayes and Chou's pseudo amino acid composition - PubMed (original) (raw)
Prediction of protein S-nitrosylation sites based on adapted normal distribution bi-profile Bayes and Chou's pseudo amino acid composition
Cangzhi Jia et al. Int J Mol Sci. 2014.
Abstract
Protein S-nitrosylation is a reversible post-translational modification by covalent modification on the thiol group of cysteine residues by nitric oxide. Growing evidence shows that protein S-nitrosylation plays an important role in normal cellular function as well as in various pathophysiologic conditions. Because of the inherent chemical instability of the S-NO bond and the low abundance of endogenous S-nitrosylated proteins, the unambiguous identification of S-nitrosylation sites by commonly used proteomic approaches remains challenging. Therefore, computational prediction of S-nitrosylation sites has been considered as a powerful auxiliary tool. In this work, we mainly adopted an adapted normal distribution bi-profile Bayes (ANBPB) feature extraction model to characterize the distinction of position-specific amino acids in 784 S-nitrosylated and 1568 non-S-nitrosylated peptide sequences. We developed a support vector machine prediction model, iSNO-ANBPB, by incorporating ANBPB with the Chou's pseudo amino acid composition. In jackknife cross-validation experiments, iSNO-ANBPB yielded an accuracy of 65.39% and a Matthew's correlation coefficient (MCC) of 0.3014. When tested on an independent dataset, iSNO-ANBPB achieved an accuracy of 63.41% and a MCC of 0.2984, which are much higher than the values achieved by the existing predictors SNOSite, iSNO-PseAAC, the Li et al. algorithm, and iSNO-AAPair. On another training dataset, iSNO-ANBPB also outperformed GPS-SNO and iSNO-PseAAC in the 10-fold crossvalidation test.
Figures
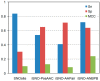
Figure 1
Potential _S_-nitrosylation sites predicted on 37 proteins through _S_-nitrosothiols (SNO)site, iSNO-PseAAC, iSNO-AAPair and iSNO-adapted normal distribution bi-profile Bayes (ANBPB) predictor.
Similar articles
- Identification of S-nitrosylation sites based on multiple features combination.
Li T, Song R, Yin Q, Gao M, Chen Y. Li T, et al. Sci Rep. 2019 Feb 28;9(1):3098. doi: 10.1038/s41598-019-39743-9. Sci Rep. 2019. PMID: 30816267 Free PMC article. - iSNO-PseAAC: predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition.
Xu Y, Ding J, Wu LY, Chou KC. Xu Y, et al. PLoS One. 2013;8(2):e55844. doi: 10.1371/journal.pone.0055844. Epub 2013 Feb 7. PLoS One. 2013. PMID: 23409062 Free PMC article. - OH-PRED: prediction of protein hydroxylation sites by incorporating adapted normal distribution bi-profile Bayes feature extraction and physicochemical properties of amino acids.
Jia CZ, He WY, Yao YH. Jia CZ, et al. J Biomol Struct Dyn. 2017 Mar;35(4):829-835. doi: 10.1080/07391102.2016.1163294. Epub 2016 May 4. J Biomol Struct Dyn. 2017. PMID: 26957000 - Computational prediction of NO-dependent posttranslational modifications in plants: Current status and perspectives.
Kolbert Z, Lindermayr C. Kolbert Z, et al. Plant Physiol Biochem. 2021 Oct;167:851-861. doi: 10.1016/j.plaphy.2021.09.011. Epub 2021 Sep 13. Plant Physiol Biochem. 2021. PMID: 34536898 Review. - Recent Advances in Predicting Protein S-Nitrosylation Sites.
Zhao Q, Ma J, Xie F, Wang Y, Zhang Y, Li H, Sun Y, Wang L, Guo M, Han K. Zhao Q, et al. Biomed Res Int. 2021 Feb 9;2021:5542224. doi: 10.1155/2021/5542224. eCollection 2021. Biomed Res Int. 2021. PMID: 33628788 Free PMC article. Review.
Cited by
- Some illuminating remarks on molecular genetics and genomics as well as drug development.
Chou KC. Chou KC. Mol Genet Genomics. 2020 Mar;295(2):261-274. doi: 10.1007/s00438-019-01634-z. Epub 2020 Jan 1. Mol Genet Genomics. 2020. PMID: 31894399 Review. - Features of reactive cysteines discovered through computation: from kinase inhibition to enrichment around protein degrons.
Fowler NJ, Blanford CF, de Visser SP, Warwicker J. Fowler NJ, et al. Sci Rep. 2017 Nov 27;7(1):16338. doi: 10.1038/s41598-017-15997-z. Sci Rep. 2017. PMID: 29180682 Free PMC article. - pCysMod: Prediction of Multiple Cysteine Modifications Based on Deep Learning Framework.
Li S, Yu K, Wu G, Zhang Q, Wang P, Zheng J, Liu ZX, Wang J, Gao X, Cheng H. Li S, et al. Front Cell Dev Biol. 2021 Feb 23;9:617366. doi: 10.3389/fcell.2021.617366. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33732693 Free PMC article. - 70ProPred: a predictor for discovering sigma70 promoters based on combining multiple features.
He W, Jia C, Duan Y, Zou Q. He W, et al. BMC Syst Biol. 2018 Apr 24;12(Suppl 4):44. doi: 10.1186/s12918-018-0570-1. BMC Syst Biol. 2018. PMID: 29745856 Free PMC article. - Sequence-based identification of recombination spots using pseudo nucleic acid representation and recursive feature extraction by linear kernel SVM.
Li L, Yu S, Xiao W, Li Y, Huang L, Zheng X, Zhou S, Yang H. Li L, et al. BMC Bioinformatics. 2014 Nov 20;15(1):340. doi: 10.1186/1471-2105-15-340. BMC Bioinformatics. 2014. PMID: 25409550 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources